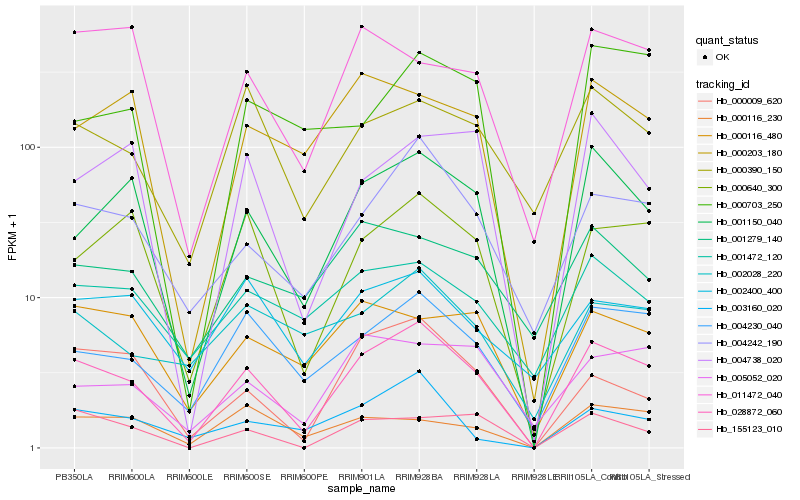
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028872_060 |
0.0 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 2 |
Hb_001150_040 |
0.1487442368 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 3 |
Hb_004242_190 |
0.1514676063 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_004230_040 |
0.1569310386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000640_300 |
0.1636448142 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 6 |
Hb_000009_620 |
0.1826624693 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_155123_010 |
0.187689764 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 8 |
Hb_000390_150 |
0.1889078554 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105632469 [Jatropha curcas] |
| 9 |
Hb_000203_180 |
0.1929342129 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10 [Jatropha curcas] |
| 10 |
Hb_005052_020 |
0.1957770026 |
- |
- |
- |
| 11 |
Hb_003160_020 |
0.1960177291 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 12 |
Hb_004738_020 |
0.1961715555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001472_120 |
0.1965765127 |
- |
- |
- |
| 14 |
Hb_002028_220 |
0.1969563101 |
- |
- |
aminoadipic semialdehyde synthase, putative [Ricinus communis] |
| 15 |
Hb_000116_480 |
0.1995625953 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 16 |
Hb_000116_230 |
0.2010489881 |
- |
- |
- |
| 17 |
Hb_000703_250 |
0.202806016 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10-like [Jatropha curcas] |
| 18 |
Hb_002400_400 |
0.2029825572 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_011472_040 |
0.2082004559 |
- |
- |
PREDICTED: clathrin light chain 3 [Jatropha curcas] |
| 20 |
Hb_001279_140 |
0.2083201286 |
- |
- |
PREDICTED: cystinosin homolog [Jatropha curcas] |