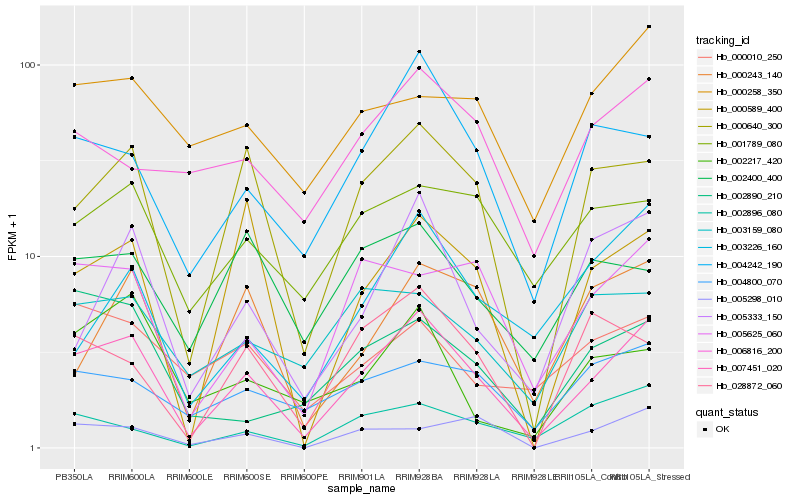
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007451_020 |
0.0 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 2 |
Hb_000640_300 |
0.1591446202 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 3 |
Hb_004242_190 |
0.1866933692 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_005625_060 |
0.1892531623 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 5 |
Hb_005333_150 |
0.1945648314 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 6 |
Hb_004800_070 |
0.204774076 |
- |
- |
- |
| 7 |
Hb_000243_140 |
0.2049384883 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 8 |
Hb_000589_400 |
0.205784353 |
- |
- |
ESC, putative [Ricinus communis] |
| 9 |
Hb_002896_080 |
0.2103193725 |
- |
- |
- |
| 10 |
Hb_003226_160 |
0.2103555851 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 11 |
Hb_001789_080 |
0.2116475432 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 12 |
Hb_006816_200 |
0.2132241139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_028872_060 |
0.2151962757 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 14 |
Hb_000258_350 |
0.217384576 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 15 |
Hb_005298_010 |
0.2177048665 |
- |
- |
- |
| 16 |
Hb_003159_080 |
0.2177177164 |
- |
- |
PREDICTED: uncharacterized protein LOC105639276 [Jatropha curcas] |
| 17 |
Hb_002890_210 |
0.2208759726 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 18 |
Hb_000010_250 |
0.2211893414 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 19 |
Hb_002217_420 |
0.2227364654 |
- |
- |
hypothetical protein EUTSA_v10021493mg [Eutrema salsugineum] |
| 20 |
Hb_002400_400 |
0.2235832161 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |