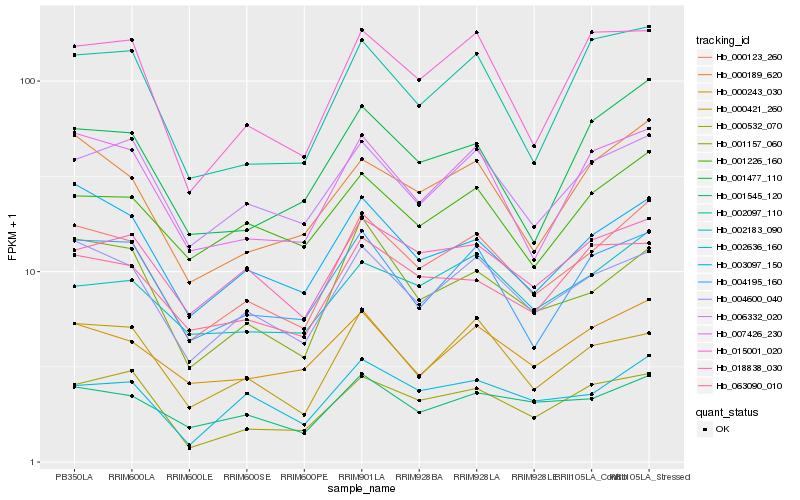
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000123_260 |
0.0 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 2 |
Hb_004600_040 |
0.0642285694 |
- |
- |
- |
| 3 |
Hb_007426_230 |
0.0762414405 |
- |
- |
hypothetical protein CICLE_v10017932mg [Citrus clementina] |
| 4 |
Hb_000189_620 |
0.0789579673 |
- |
- |
hypothetical protein JCGZ_23450 [Jatropha curcas] |
| 5 |
Hb_001545_120 |
0.0810254071 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g34160 [Jatropha curcas] |
| 6 |
Hb_003097_150 |
0.0826490095 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 7 |
Hb_004195_160 |
0.083909878 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 8 |
Hb_006332_020 |
0.0853595088 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 9 |
Hb_000243_030 |
0.0863335599 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 10 |
Hb_001157_060 |
0.0885865293 |
- |
- |
PREDICTED: putative nitric oxide synthase [Jatropha curcas] |
| 11 |
Hb_001226_160 |
0.0897756915 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein B''-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000532_070 |
0.0900838133 |
- |
- |
PREDICTED: exosome complex component RRP42-like [Jatropha curcas] |
| 13 |
Hb_015001_020 |
0.090696895 |
- |
- |
PREDICTED: la-related protein 6B [Jatropha curcas] |
| 14 |
Hb_002097_110 |
0.0910710652 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002183_090 |
0.0914700155 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 16 |
Hb_002636_160 |
0.0917359882 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g13630 [Jatropha curcas] |
| 17 |
Hb_018838_030 |
0.0924118786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000421_260 |
0.0929822489 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 19 |
Hb_063090_010 |
0.093125695 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001477_110 |
0.0931490511 |
- |
- |
prohibitin, putative [Ricinus communis] |