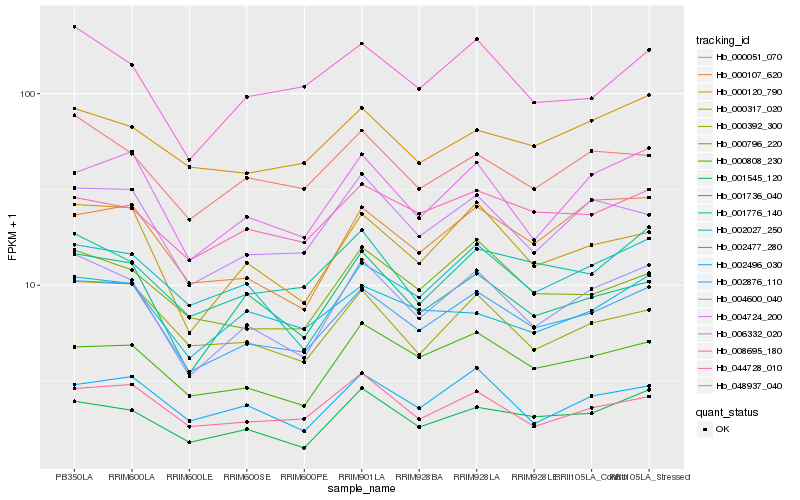
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000317_020 |
0.0605205267 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 3 |
Hb_004600_040 |
0.0617116553 |
- |
- |
- |
| 4 |
Hb_006332_020 |
0.0690008326 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 5 |
Hb_004724_200 |
0.0700063792 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 6 |
Hb_001736_040 |
0.0717227168 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 7 |
Hb_000392_300 |
0.0722394386 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 8 |
Hb_048937_040 |
0.0749849168 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_000808_230 |
0.0752882243 |
- |
- |
PREDICTED: peptide chain release factor PrfB2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001545_120 |
0.0781061497 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g34160 [Jatropha curcas] |
| 11 |
Hb_000107_620 |
0.0803704135 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 12 |
Hb_001776_140 |
0.0806096696 |
- |
- |
PREDICTED: probable GTP-binding protein OBGM, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_002477_280 |
0.0807954034 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-2 [Jatropha curcas] |
| 14 |
Hb_008695_180 |
0.0810761001 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 15 |
Hb_002496_030 |
0.0814334984 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g69350, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002027_250 |
0.0814545255 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000051_070 |
0.0817707816 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |
| 18 |
Hb_000796_220 |
0.0821240972 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000120_790 |
0.0832625095 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 20 |
Hb_044728_010 |
0.0837257631 |
- |
- |
- |