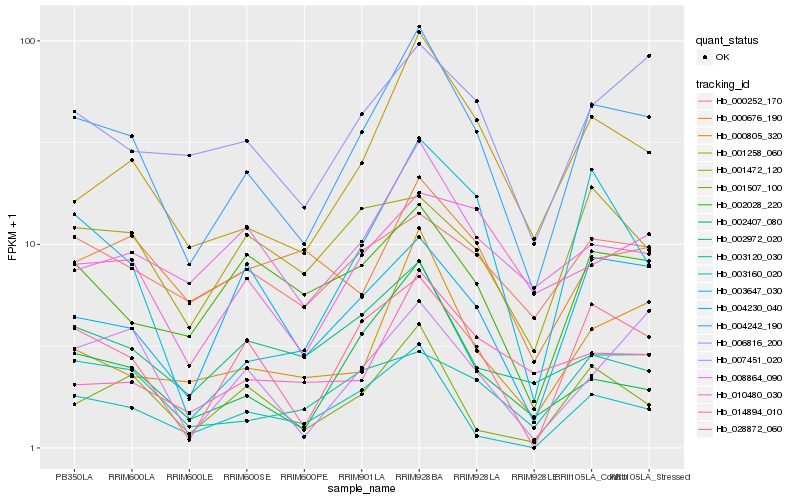
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004242_190 |
0.0 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 2 |
Hb_001258_060 |
0.1332862542 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 3 |
Hb_028872_060 |
0.1514676063 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 4 |
Hb_000805_320 |
0.1568247703 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 5 |
Hb_006816_200 |
0.1582662863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002407_080 |
0.1624870201 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 7 |
Hb_003120_030 |
0.1648615675 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 8 |
Hb_002028_220 |
0.1693420095 |
- |
- |
aminoadipic semialdehyde synthase, putative [Ricinus communis] |
| 9 |
Hb_008864_090 |
0.1724716074 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 10 |
Hb_004230_040 |
0.1751867795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010480_030 |
0.1759734195 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_003160_020 |
0.176805666 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 13 |
Hb_003647_030 |
0.1771808798 |
- |
- |
PREDICTED: probable glutathione S-transferase [Populus euphratica] |
| 14 |
Hb_002972_020 |
0.1788240024 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 15 |
Hb_000252_170 |
0.1803682565 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001507_100 |
0.1840951432 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_014894_010 |
0.1865054675 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 18 |
Hb_007451_020 |
0.1866933692 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 19 |
Hb_000676_190 |
0.1885450364 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |
| 20 |
Hb_001472_120 |
0.1886146604 |
- |
- |
- |