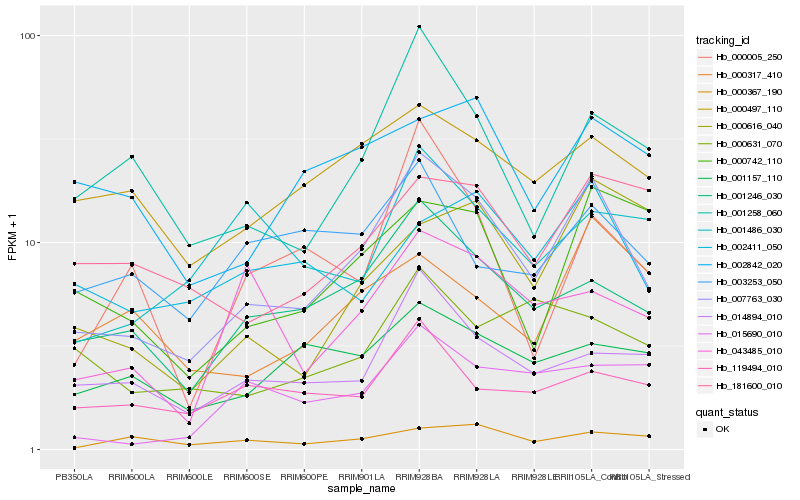
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007763_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 2 |
Hb_001246_030 |
0.1535277319 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 3 |
Hb_001258_060 |
0.1765023658 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 4 |
Hb_014894_010 |
0.1826776982 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 5 |
Hb_000497_110 |
0.1897143653 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 6 |
Hb_000742_110 |
0.1950012225 |
- |
- |
hypothetical protein JCGZ_09215 [Jatropha curcas] |
| 7 |
Hb_181600_010 |
0.2028008335 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 8 |
Hb_001157_110 |
0.203760111 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 9 |
Hb_002842_020 |
0.2064205534 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 10 |
Hb_043485_010 |
0.2068384253 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_119494_010 |
0.2069446884 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 12 |
Hb_000005_250 |
0.2080123244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002411_050 |
0.2090068958 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000367_190 |
0.2111633275 |
- |
- |
hypothetical protein RCOM_1470550 [Ricinus communis] |
| 15 |
Hb_015690_010 |
0.2115492821 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g038521mg [Citrus sinensis] |
| 16 |
Hb_000631_070 |
0.2128700982 |
transcription factor |
TF Family: C2H2 |
PREDICTED: histone deacetylase 5 [Jatropha curcas] |
| 17 |
Hb_000317_410 |
0.2147037821 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 18 |
Hb_003253_050 |
0.2159247209 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 19 |
Hb_000616_040 |
0.2165787393 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 20 |
Hb_001486_030 |
0.2170919285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |