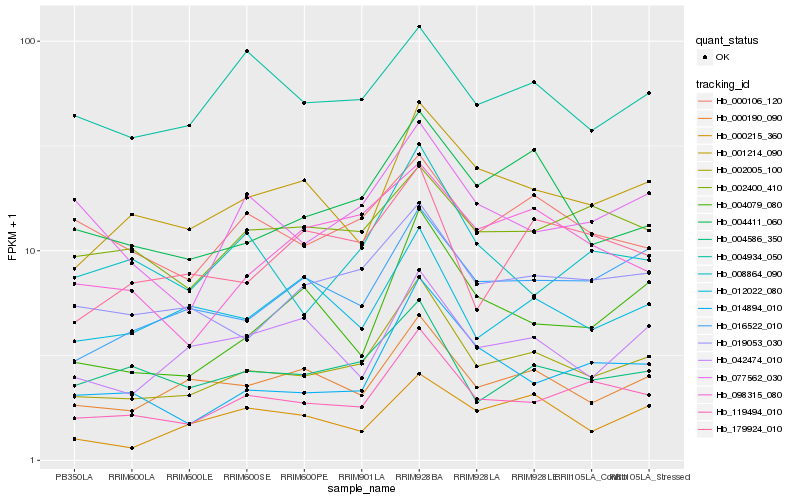
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_119494_010 |
0.0926497892 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 3 |
Hb_000190_090 |
0.105983733 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 4 |
Hb_019053_030 |
0.1092203158 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 5 |
Hb_004411_060 |
0.1106968358 |
- |
- |
acetolactate synthase, putative [Ricinus communis] |
| 6 |
Hb_001214_090 |
0.1151099519 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 7 |
Hb_016522_010 |
0.1174630155 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 8 |
Hb_004586_350 |
0.1214353549 |
- |
- |
Phosphoribosylaminoimidazole-succinocarboxamide synthase [Morus notabilis] |
| 9 |
Hb_014894_010 |
0.1233078868 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 10 |
Hb_004079_080 |
0.1280098472 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 11 |
Hb_077562_030 |
0.1314155303 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_012022_080 |
0.1331874042 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000106_120 |
0.1353739761 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 14 |
Hb_179924_010 |
0.1356223455 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_008864_090 |
0.1356841024 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 16 |
Hb_042474_010 |
0.1358193728 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 17 |
Hb_098315_080 |
0.1361721891 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_002400_410 |
0.1385690911 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 19 |
Hb_000215_360 |
0.1386424247 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 20 |
Hb_004934_050 |
0.1396584469 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |