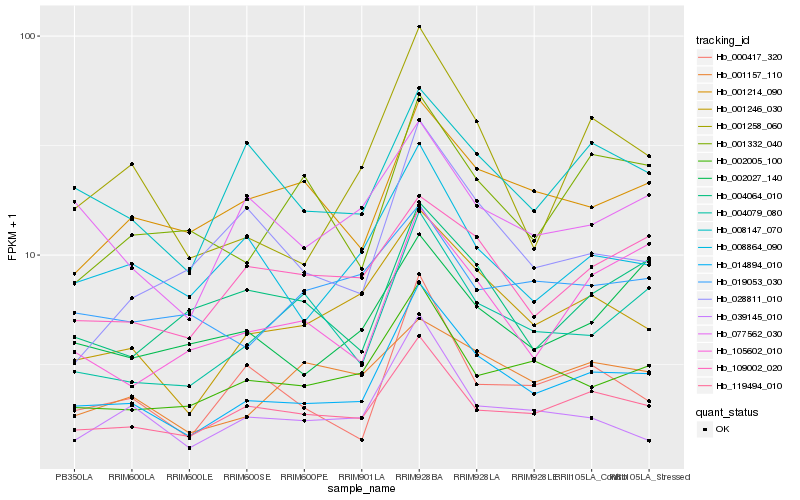
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014894_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 2 |
Hb_001246_030 |
0.100147261 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 3 |
Hb_119494_010 |
0.1033266389 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 4 |
Hb_002005_100 |
0.1233078868 |
- |
- |
- |
| 5 |
Hb_001258_060 |
0.1254735988 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 6 |
Hb_004079_080 |
0.1264893348 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 7 |
Hb_008864_090 |
0.1326456444 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 8 |
Hb_008147_070 |
0.1370832489 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 9 |
Hb_001214_090 |
0.1396362482 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 10 |
Hb_000417_320 |
0.1406692505 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 11 |
Hb_109002_020 |
0.1420311131 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 12 |
Hb_039145_010 |
0.1427168601 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 13 |
Hb_077562_030 |
0.1430065929 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001157_110 |
0.1479363282 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 15 |
Hb_004064_010 |
0.1508313136 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 16 |
Hb_105602_010 |
0.1510337725 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 17 |
Hb_001332_040 |
0.1532826656 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 18 |
Hb_002027_140 |
0.1576343716 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 19 |
Hb_028811_010 |
0.1579080283 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 20 |
Hb_019053_030 |
0.1593621447 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |