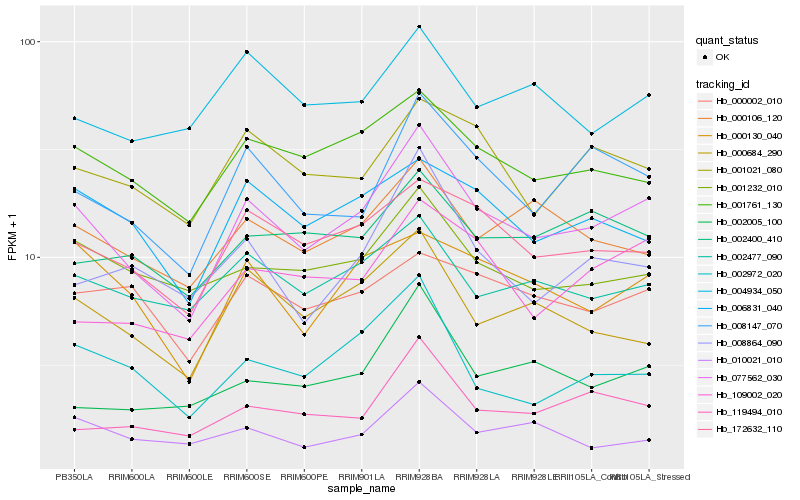
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077562_030 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_008147_070 |
0.1030013054 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 3 |
Hb_172632_110 |
0.1075406978 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 4 |
Hb_001761_130 |
0.1119577699 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002477_090 |
0.1198852563 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001232_010 |
0.1199378753 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 7 |
Hb_000684_290 |
0.1204439418 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 8 |
Hb_000106_120 |
0.1220876665 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 9 |
Hb_109002_020 |
0.1234900919 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 10 |
Hb_119494_010 |
0.1240909962 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 11 |
Hb_000130_040 |
0.1243748566 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 12 |
Hb_001021_080 |
0.1247948353 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 13 |
Hb_006831_040 |
0.1258932119 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002972_020 |
0.1269515483 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 15 |
Hb_008864_090 |
0.1278544599 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 16 |
Hb_002400_410 |
0.1297705084 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 17 |
Hb_002005_100 |
0.1314155303 |
- |
- |
- |
| 18 |
Hb_010021_010 |
0.1334679488 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_004934_050 |
0.1351265661 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 20 |
Hb_000002_010 |
0.1353081075 |
- |
- |
PREDICTED: uncharacterized protein LOC105641567 [Jatropha curcas] |