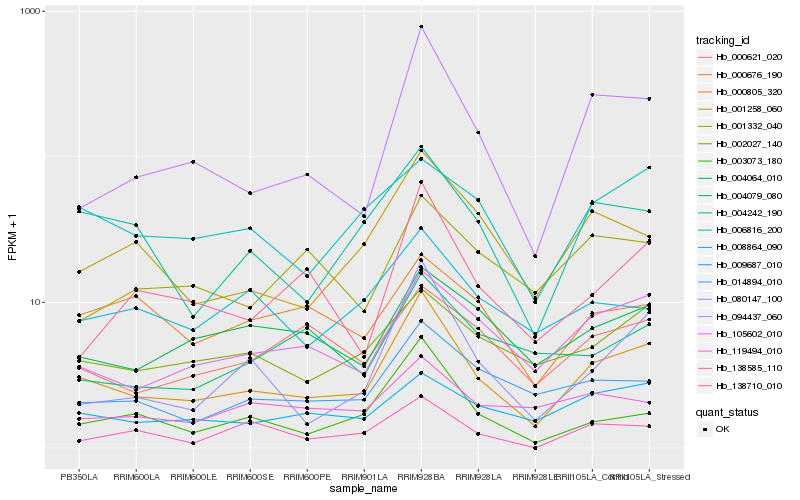
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_320 |
0.0 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 2 |
Hb_094437_060 |
0.1420891572 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 3 |
Hb_004242_190 |
0.1568247703 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_105602_010 |
0.156892455 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 5 |
Hb_001258_060 |
0.1648023413 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 6 |
Hb_003073_180 |
0.168723814 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 7 |
Hb_014894_010 |
0.1691370523 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 8 |
Hb_080147_100 |
0.1699687746 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 9 |
Hb_009687_010 |
0.1766973026 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 10 |
Hb_002027_140 |
0.176951975 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 11 |
Hb_138710_010 |
0.1775717683 |
- |
- |
Adenosylmethionine-8-amino-7-oxononanoate aminotransferase [Gossypium arboreum] |
| 12 |
Hb_006816_200 |
0.1826418755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004064_010 |
0.1842124669 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 14 |
Hb_000621_020 |
0.1905119309 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 15 |
Hb_008864_090 |
0.191261605 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 16 |
Hb_119494_010 |
0.1956814815 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 17 |
Hb_138585_110 |
0.2017284088 |
- |
- |
- |
| 18 |
Hb_004079_080 |
0.201748659 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 19 |
Hb_001332_040 |
0.2035612004 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000676_190 |
0.2041482057 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |