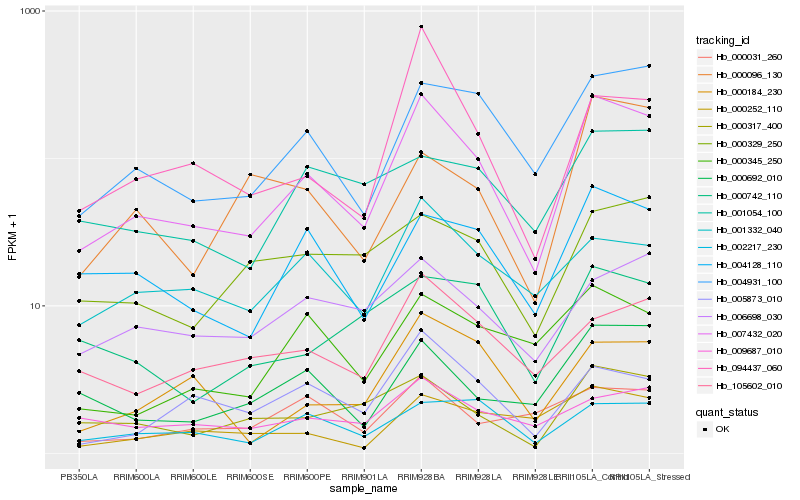
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007432_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s13310g [Populus trichocarpa] |
| 2 |
Hb_004931_100 |
0.1433747454 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 3 |
Hb_004128_110 |
0.1588241345 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 4 |
Hb_000317_400 |
0.1633606446 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000692_010 |
0.168440031 |
- |
- |
PREDICTED: uncharacterized protein LOC105635295 [Jatropha curcas] |
| 6 |
Hb_105602_010 |
0.1769628505 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 7 |
Hb_002217_230 |
0.1813652204 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 8 |
Hb_000184_230 |
0.1815309942 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 9-like [Jatropha curcas] |
| 9 |
Hb_005873_010 |
0.1815927035 |
- |
- |
cysteine desulfurylase, putative [Ricinus communis] |
| 10 |
Hb_001332_040 |
0.1827714057 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 11 |
Hb_094437_060 |
0.185575022 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 12 |
Hb_000345_250 |
0.1858209294 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 13 |
Hb_000329_250 |
0.1865488073 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_006698_030 |
0.1872479935 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 15 |
Hb_000252_110 |
0.1904292468 |
- |
- |
- |
| 16 |
Hb_000742_110 |
0.1910393065 |
- |
- |
hypothetical protein JCGZ_09215 [Jatropha curcas] |
| 17 |
Hb_001054_100 |
0.1937620475 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 18 |
Hb_000096_130 |
0.1952786361 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 19 |
Hb_009687_010 |
0.1983290754 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 20 |
Hb_000031_260 |
0.1985064129 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |