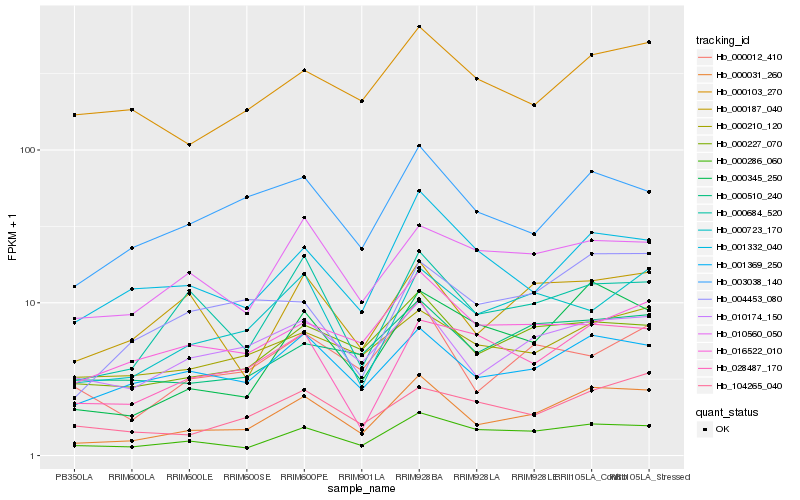
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_260 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 2 |
Hb_000286_060 |
0.1206020703 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 3 |
Hb_000345_250 |
0.120736752 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 4 |
Hb_000227_070 |
0.1261719482 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 5 |
Hb_001369_250 |
0.1269987753 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010174_150 |
0.1287799477 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_003038_140 |
0.1389532283 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 8 |
Hb_000684_520 |
0.1407339564 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 9 |
Hb_001332_040 |
0.1415247716 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000510_240 |
0.1418744822 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000723_170 |
0.1419301151 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 12 |
Hb_000012_410 |
0.1449566936 |
- |
- |
PREDICTED: histone deacetylase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_104265_040 |
0.1482772572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004453_080 |
0.1483746178 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 15 |
Hb_028487_170 |
0.1498823318 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 16 |
Hb_000210_120 |
0.1508425259 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000103_270 |
0.1521165636 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_010560_050 |
0.152731749 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 19 |
Hb_016522_010 |
0.1531612886 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 20 |
Hb_000187_040 |
0.1535208848 |
- |
- |
PREDICTED: pyridoxal kinase isoform X5 [Vitis vinifera] |