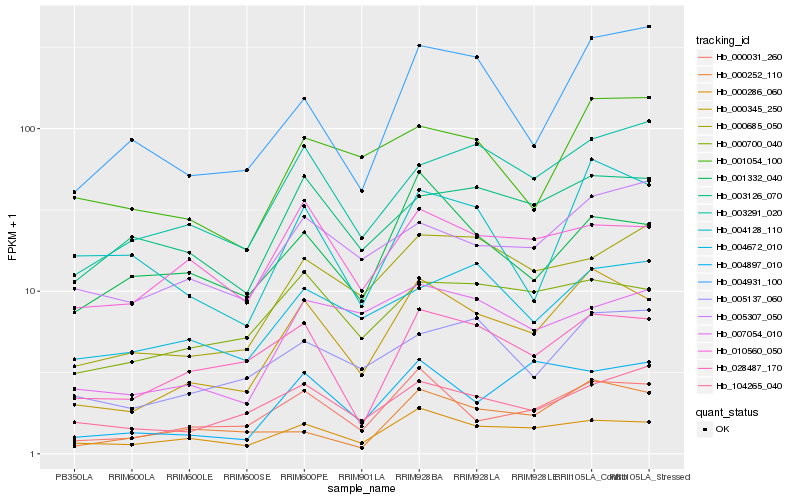
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_250 |
0.0 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 2 |
Hb_000031_260 |
0.120736752 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 3 |
Hb_000286_060 |
0.1291467777 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 4 |
Hb_028487_170 |
0.1528758419 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 5 |
Hb_003291_020 |
0.1564135219 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 6 |
Hb_000700_040 |
0.1584898937 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |
| 7 |
Hb_000685_050 |
0.1601262843 |
- |
- |
PREDICTED: mitochondrial inner membrane protein OXA1-like [Populus euphratica] |
| 8 |
Hb_005137_060 |
0.1618745069 |
- |
- |
- |
| 9 |
Hb_004931_100 |
0.1671460487 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 10 |
Hb_004897_010 |
0.1671498093 |
- |
- |
- |
| 11 |
Hb_010560_050 |
0.1705286065 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 12 |
Hb_007054_010 |
0.1708949021 |
- |
- |
PREDICTED: WD repeat-containing protein 82 [Jatropha curcas] |
| 13 |
Hb_005307_050 |
0.1718597989 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |
| 14 |
Hb_104265_040 |
0.1724441991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003126_070 |
0.1726121702 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001332_040 |
0.1744286207 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 17 |
Hb_004128_110 |
0.175670849 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 18 |
Hb_000252_110 |
0.1758782639 |
- |
- |
- |
| 19 |
Hb_004672_010 |
0.1760930565 |
- |
- |
ecotropic viral integration site, putative [Ricinus communis] |
| 20 |
Hb_001054_100 |
0.1765813851 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |