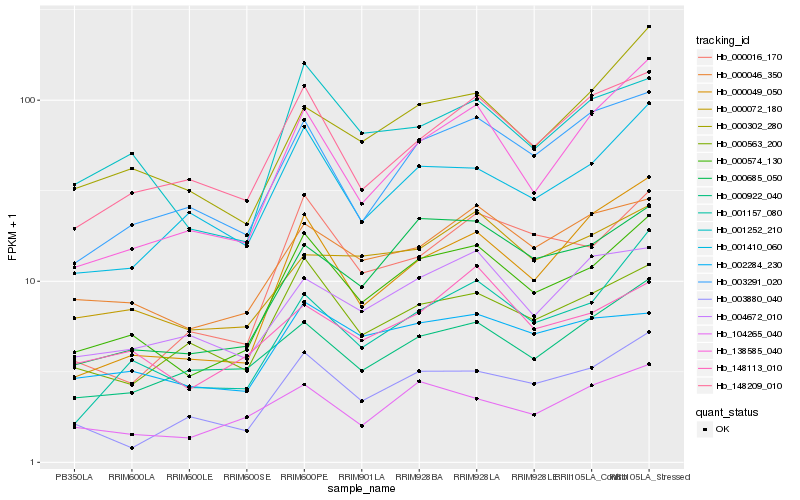
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_130 |
0.0 |
- |
- |
calcineurin-like protein [Hevea brasiliensis] |
| 2 |
Hb_000046_350 |
0.1001339042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000685_050 |
0.1012631102 |
- |
- |
PREDICTED: mitochondrial inner membrane protein OXA1-like [Populus euphratica] |
| 4 |
Hb_001252_210 |
0.102646912 |
- |
- |
PREDICTED: glycine cleavage system H protein 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000563_200 |
0.1072991381 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |
| 6 |
Hb_148209_010 |
0.1077327793 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 7 |
Hb_003880_040 |
0.1089480355 |
- |
- |
hypothetical protein POPTR_0001s30770g [Populus trichocarpa] |
| 8 |
Hb_001410_060 |
0.1102527237 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 9 |
Hb_003291_020 |
0.1105034682 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 10 |
Hb_001157_080 |
0.11342551 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_138585_040 |
0.1142963901 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 12 |
Hb_104265_040 |
0.114749907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000302_280 |
0.1183314445 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 14 |
Hb_000072_180 |
0.1190025576 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |
| 15 |
Hb_000016_170 |
0.1195209991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004672_010 |
0.1196080246 |
- |
- |
ecotropic viral integration site, putative [Ricinus communis] |
| 17 |
Hb_002284_230 |
0.1205586927 |
- |
- |
PREDICTED: uncharacterized protein At5g43822 [Jatropha curcas] |
| 18 |
Hb_000922_040 |
0.1217021513 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 19 |
Hb_000049_050 |
0.1221005081 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 20 |
Hb_148113_010 |
0.1227062456 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17C-like isoform X3 [Citrus sinensis] |