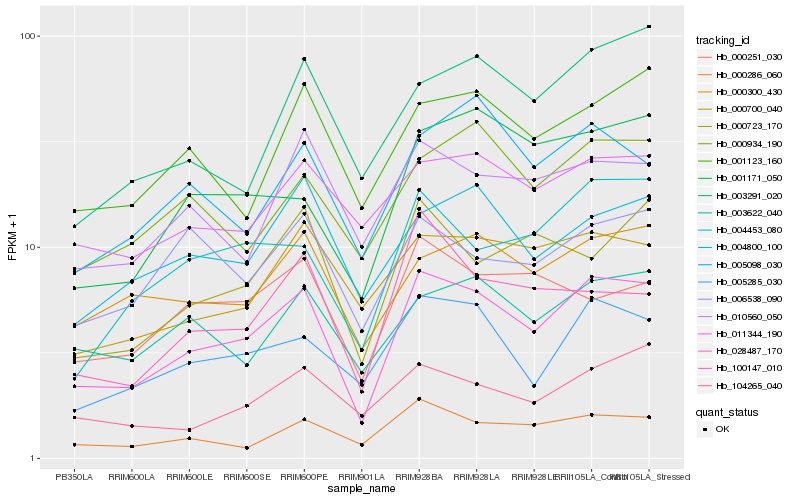
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028487_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 2 |
Hb_005285_030 |
0.1224290874 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 3 |
Hb_000286_060 |
0.1262719072 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 4 |
Hb_000300_430 |
0.1270136186 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 5 |
Hb_001123_160 |
0.1319060288 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 6 |
Hb_000700_040 |
0.1329906927 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |
| 7 |
Hb_004453_080 |
0.1332599455 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 8 |
Hb_003622_040 |
0.1337287271 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 9 |
Hb_004800_100 |
0.1343892012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000934_190 |
0.1356103003 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 11 |
Hb_000251_030 |
0.1359295515 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000723_170 |
0.1362057078 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 13 |
Hb_104265_040 |
0.1366484803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010560_050 |
0.1368072048 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 15 |
Hb_011344_190 |
0.1371756849 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 16 |
Hb_001171_050 |
0.1395445081 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 17 |
Hb_005098_030 |
0.1412417606 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006538_090 |
0.1419690714 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 19 |
Hb_003291_020 |
0.1419945929 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 20 |
Hb_100147_010 |
0.1432125168 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |