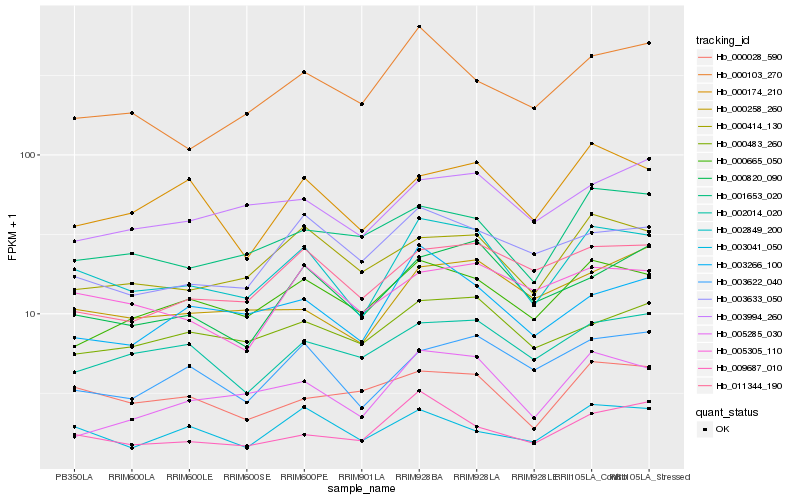
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002849_200 |
0.0 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 2 |
Hb_000414_130 |
0.1080060483 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 3 |
Hb_000665_050 |
0.1105956258 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 4 |
Hb_003622_040 |
0.1132197316 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 5 |
Hb_003266_100 |
0.1147604549 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 6 |
Hb_000820_090 |
0.1149304681 |
- |
- |
- |
| 7 |
Hb_000258_260 |
0.1177313201 |
- |
- |
PREDICTED: protein HGH1 homolog [Jatropha curcas] |
| 8 |
Hb_001653_020 |
0.117784254 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 9 |
Hb_000483_260 |
0.1188945789 |
- |
- |
PREDICTED: dihydroorotase, mitochondrial isoform X1 [Populus euphratica] |
| 10 |
Hb_009687_010 |
0.1191166611 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 11 |
Hb_000174_210 |
0.1191289318 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 12 |
Hb_011344_190 |
0.1196236661 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 13 |
Hb_005285_030 |
0.1202068674 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 14 |
Hb_005305_110 |
0.1203706705 |
- |
- |
PREDICTED: uncharacterized protein LOC105648286 [Jatropha curcas] |
| 15 |
Hb_003994_260 |
0.1205154647 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 16 |
Hb_002014_020 |
0.1208116964 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000028_590 |
0.1216249389 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 18 |
Hb_003633_050 |
0.1222343649 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 19 |
Hb_000103_270 |
0.1222602015 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_003041_050 |
0.1229308767 |
- |
- |
- |