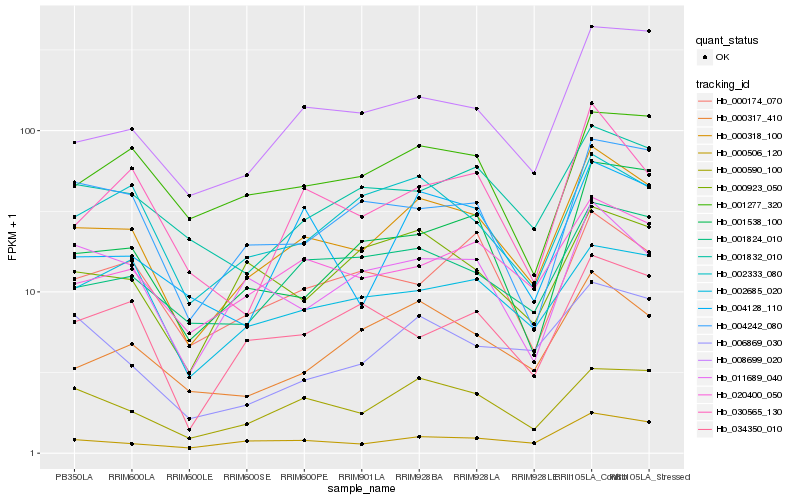
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_100 |
0.0 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 2 |
Hb_000590_100 |
0.1084737807 |
- |
- |
- |
| 3 |
Hb_020400_050 |
0.1114158642 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 4 |
Hb_000506_120 |
0.114053835 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 5 |
Hb_001824_010 |
0.1172578073 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 6 |
Hb_000317_410 |
0.1269670613 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 7 |
Hb_008699_020 |
0.1270286475 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 8 |
Hb_001832_010 |
0.1308671439 |
- |
- |
PREDICTED: uncharacterized protein LOC105637890 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002333_080 |
0.1346705476 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20-like [Populus euphratica] |
| 10 |
Hb_011689_040 |
0.1348015473 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 11 |
Hb_001277_320 |
0.1350234489 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 12 |
Hb_004242_080 |
0.1358281438 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 13 |
Hb_006869_030 |
0.13734126 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_002685_020 |
0.1376875775 |
- |
- |
- |
| 15 |
Hb_030565_130 |
0.1381161485 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 16 |
Hb_004128_110 |
0.1385392023 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 17 |
Hb_001538_100 |
0.1390046798 |
- |
- |
PREDICTED: histone deacetylase 6 [Jatropha curcas] |
| 18 |
Hb_000923_050 |
0.1406144024 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 19 |
Hb_034350_010 |
0.1407415724 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Populus euphratica] |
| 20 |
Hb_000174_070 |
0.1427105485 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |