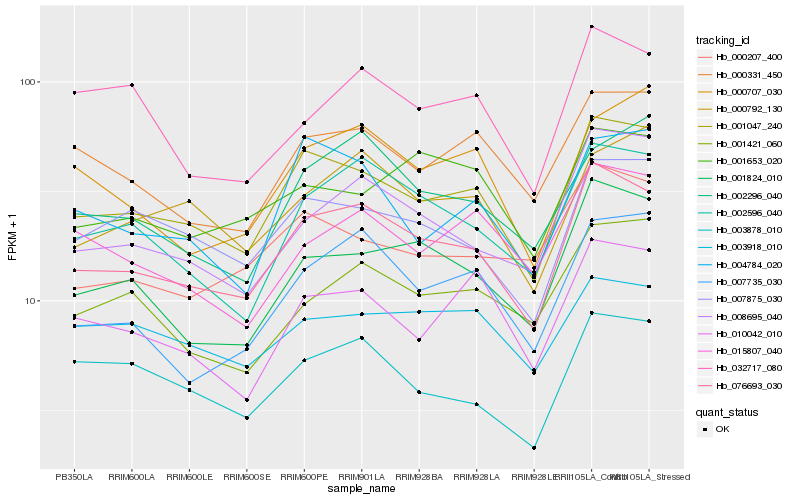
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_076693_030 |
0.0 |
- |
- |
PREDICTED: serine/arginine-rich-splicing factor SR34-like [Populus euphratica] |
| 2 |
Hb_001047_240 |
0.0546971088 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 20-like [Jatropha curcas] |
| 3 |
Hb_002596_040 |
0.0757215721 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001824_010 |
0.0842314648 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 5 |
Hb_008695_040 |
0.0850631448 |
- |
- |
PREDICTED: small ribosomal subunit protein S13, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_007875_030 |
0.0937260077 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 7 |
Hb_007735_030 |
0.0961604949 |
- |
- |
Calcium-binding EF-hand family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_000331_450 |
0.0981673722 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 6 homolog [Jatropha curcas] |
| 9 |
Hb_003878_010 |
0.1020995463 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 10 |
Hb_002296_040 |
0.1054524254 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 11 |
Hb_000792_130 |
0.1056190104 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 12 |
Hb_000207_400 |
0.1066343468 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase PTP1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001421_060 |
0.107912476 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2B isoform X2 [Jatropha curcas] |
| 14 |
Hb_000707_030 |
0.1085792139 |
- |
- |
PREDICTED: probable proteasome inhibitor [Vitis vinifera] |
| 15 |
Hb_003918_010 |
0.1086197187 |
- |
- |
PREDICTED: biotin--protein ligase 2-like [Jatropha curcas] |
| 16 |
Hb_032717_080 |
0.1087006364 |
- |
- |
gamma-soluble nsf attachment protein, putative [Ricinus communis] |
| 17 |
Hb_015807_040 |
0.1088605526 |
- |
- |
PREDICTED: polycomb group protein FERTILIZATION-INDEPENDENT ENDOSPERM [Jatropha curcas] |
| 18 |
Hb_010042_010 |
0.1096064745 |
- |
- |
Urease accessory protein ureD, putative [Ricinus communis] |
| 19 |
Hb_001653_020 |
0.1098597581 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 20 |
Hb_004784_020 |
0.1100280252 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |