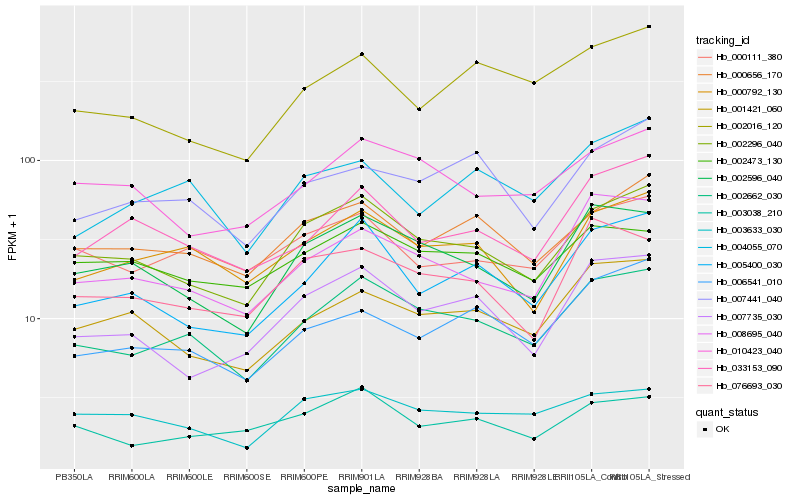
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002662_030 |
0.0 |
- |
- |
PREDICTED: mannose-P-dolichol utilization defect 1 protein homolog 2 [Jatropha curcas] |
| 2 |
Hb_002296_040 |
0.0770969077 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 3 |
Hb_002596_040 |
0.0864189528 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000792_130 |
0.0889253783 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 5 |
Hb_008695_040 |
0.0943626029 |
- |
- |
PREDICTED: small ribosomal subunit protein S13, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_005400_030 |
0.0978531486 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 7 |
Hb_000656_170 |
0.1032669787 |
- |
- |
lsm1, putative [Ricinus communis] |
| 8 |
Hb_001421_060 |
0.1036088674 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2B isoform X2 [Jatropha curcas] |
| 9 |
Hb_007735_030 |
0.1056593883 |
- |
- |
Calcium-binding EF-hand family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_010423_040 |
0.105950046 |
- |
- |
hypothetical protein JCGZ_17463 [Jatropha curcas] |
| 11 |
Hb_003038_210 |
0.1077299037 |
- |
- |
hypothetical protein B456_006G076400 [Gossypium raimondii] |
| 12 |
Hb_000111_380 |
0.1086199427 |
- |
- |
PREDICTED: cilia- and flagella-associated protein 20 [Jatropha curcas] |
| 13 |
Hb_006541_010 |
0.1088664595 |
- |
- |
PREDICTED: uncharacterized protein LOC105637261 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007441_040 |
0.1100900373 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 15 |
Hb_076693_030 |
0.1107837701 |
- |
- |
PREDICTED: serine/arginine-rich-splicing factor SR34-like [Populus euphratica] |
| 16 |
Hb_033153_090 |
0.1120063973 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 17 |
Hb_002473_130 |
0.1140422088 |
- |
- |
PREDICTED: THO complex subunit 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002016_120 |
0.114550976 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940 [Jatropha curcas] |
| 19 |
Hb_003633_030 |
0.115374204 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 20 |
Hb_004055_070 |
0.115878702 |
- |
- |
PREDICTED: uncharacterized protein LOC105631439 isoform X2 [Jatropha curcas] |