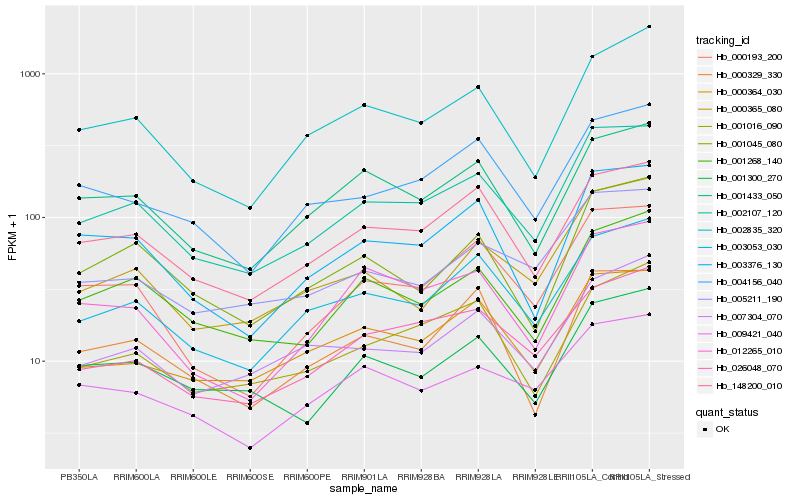
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002107_120 |
0.0 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 2 |
Hb_003053_030 |
0.0734880344 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 3 |
Hb_148200_010 |
0.0757933735 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 4 |
Hb_000193_200 |
0.0764300976 |
- |
- |
rotamase, putative [Ricinus communis] |
| 5 |
Hb_003376_130 |
0.0787279505 |
transcription factor |
TF Family: S1Fa-like |
DNA-binding protein S1FA, putative [Ricinus communis] |
| 6 |
Hb_001268_140 |
0.082024902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005211_190 |
0.0828118044 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 8 |
Hb_000364_030 |
0.0853597853 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002835_320 |
0.0855754984 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 10 |
Hb_004156_040 |
0.0860129452 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 11 |
Hb_001045_080 |
0.0873775923 |
- |
- |
PREDICTED: uncharacterized protein LOC105646704 [Jatropha curcas] |
| 12 |
Hb_001300_270 |
0.088940959 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 13 |
Hb_009421_040 |
0.0891609069 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 14 |
Hb_026048_070 |
0.0896129753 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 15 |
Hb_000329_330 |
0.0915752167 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 16 |
Hb_001433_050 |
0.093656198 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 17 |
Hb_000365_080 |
0.0937919921 |
- |
- |
PREDICTED: uncharacterized protein LOC105649037 [Jatropha curcas] |
| 18 |
Hb_012265_010 |
0.0941691527 |
- |
- |
hypothetical protein CISIN_1g026883mg [Citrus sinensis] |
| 19 |
Hb_001016_090 |
0.0942629924 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 1 [Populus euphratica] |
| 20 |
Hb_007304_070 |
0.0943969369 |
- |
- |
hypoxanthine-guanine phosphoribosyltransferase, putative [Ricinus communis] |