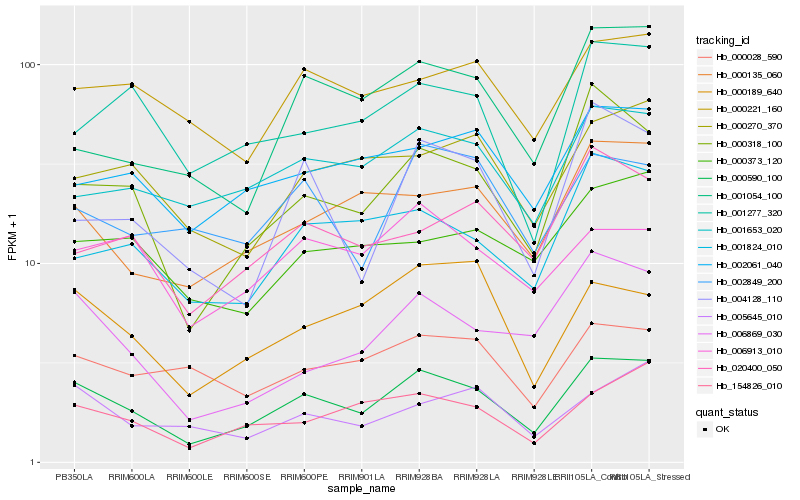
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000590_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_000318_100 |
0.1084737807 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 3 |
Hb_154826_010 |
0.1240840812 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_000135_060 |
0.1245105813 |
transcription factor |
TF Family: MED6 |
PREDICTED: mediator of RNA polymerase II transcription subunit 6 [Jatropha curcas] |
| 5 |
Hb_001824_010 |
0.1311443436 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 6 |
Hb_001653_020 |
0.1342253405 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 7 |
Hb_002061_040 |
0.134817376 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 8 |
Hb_000270_370 |
0.1364745424 |
- |
- |
Uncharacterized protein TCM_011954 [Theobroma cacao] |
| 9 |
Hb_000189_640 |
0.1380435217 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 10 |
Hb_000221_160 |
0.1404096084 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6b-1 [Jatropha curcas] |
| 11 |
Hb_006869_030 |
0.1409745638 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_001277_320 |
0.1413662878 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 13 |
Hb_001054_100 |
0.1422197794 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 14 |
Hb_005645_010 |
0.1424587108 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |
| 15 |
Hb_004128_110 |
0.142576873 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 16 |
Hb_000373_120 |
0.1429536441 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 17 |
Hb_020400_050 |
0.1436686804 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000028_590 |
0.1443712231 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 19 |
Hb_006913_010 |
0.1445489335 |
- |
- |
PREDICTED: uncharacterized protein LOC105649144 [Jatropha curcas] |
| 20 |
Hb_002849_200 |
0.1451865543 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |