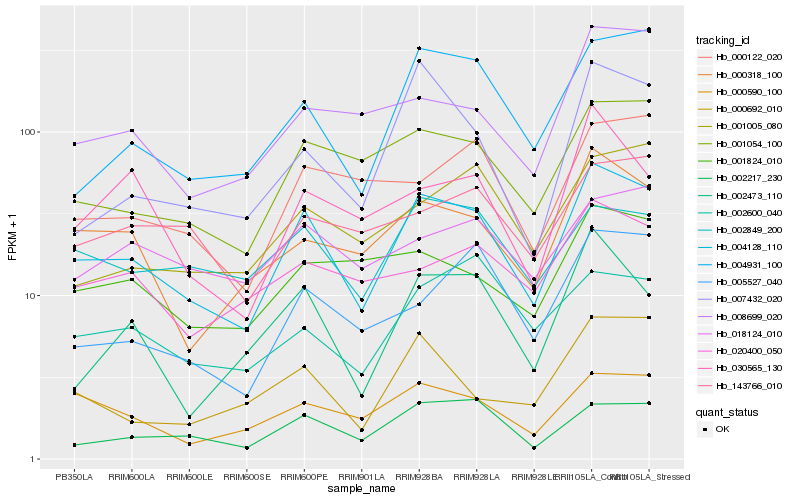
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004128_110 |
0.0 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 2 |
Hb_001054_100 |
0.1312439585 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 3 |
Hb_002217_230 |
0.1313386307 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 4 |
Hb_004931_100 |
0.1347587602 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000318_100 |
0.1385392023 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 6 |
Hb_000590_100 |
0.142576873 |
- |
- |
- |
| 7 |
Hb_002849_200 |
0.1452788033 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 8 |
Hb_002473_110 |
0.1516830482 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_007432_020 |
0.1588241345 |
- |
- |
hypothetical protein POPTR_0010s13310g [Populus trichocarpa] |
| 10 |
Hb_005527_040 |
0.1589585988 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 11 |
Hb_001824_010 |
0.1594411858 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 12 |
Hb_020400_050 |
0.1611760609 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 13 |
Hb_008699_020 |
0.161974758 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 14 |
Hb_000122_020 |
0.1630920663 |
- |
- |
PREDICTED: alpha-soluble NSF attachment protein 2 [Jatropha curcas] |
| 15 |
Hb_000692_010 |
0.1632898879 |
- |
- |
PREDICTED: uncharacterized protein LOC105635295 [Jatropha curcas] |
| 16 |
Hb_001005_080 |
0.1647117032 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_143766_010 |
0.1649064971 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 18 |
Hb_002600_040 |
0.1650294237 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 19 |
Hb_018124_010 |
0.1654242634 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_030565_130 |
0.1666175063 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |