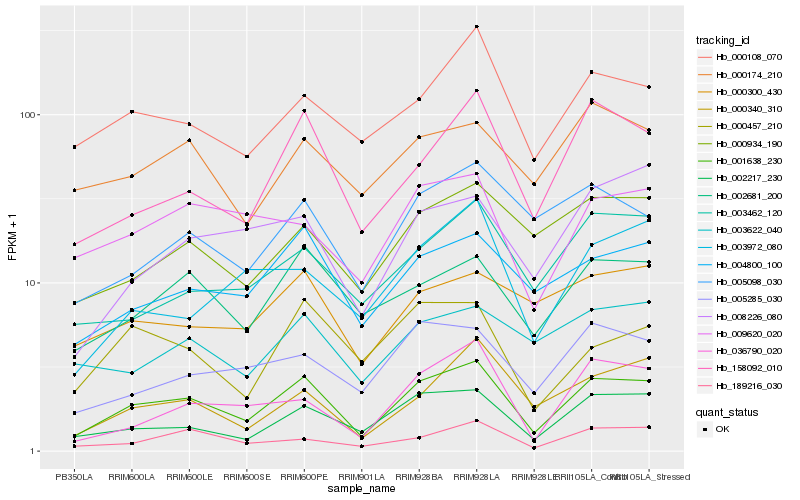
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_230 |
0.0 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 2 |
Hb_002217_230 |
0.1241548495 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 3 |
Hb_158092_010 |
0.1358292615 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 4 |
Hb_004800_100 |
0.1388032094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002681_200 |
0.1399206595 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 6 |
Hb_000108_070 |
0.1495768881 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 7 |
Hb_189216_030 |
0.1515308016 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 8 |
Hb_005285_030 |
0.1516099818 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 9 |
Hb_000934_190 |
0.154009673 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 10 |
Hb_009620_020 |
0.1551436823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000340_310 |
0.1555233965 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 12 |
Hb_005098_030 |
0.1562323218 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003462_120 |
0.1571088874 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 14 |
Hb_000457_210 |
0.1577369531 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 15 |
Hb_036790_020 |
0.1629864836 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 16 |
Hb_008226_080 |
0.1633834075 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 17 |
Hb_000174_210 |
0.1645184674 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 18 |
Hb_000300_430 |
0.1647135623 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 19 |
Hb_003622_040 |
0.1648176391 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 20 |
Hb_003972_080 |
0.1652533154 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |