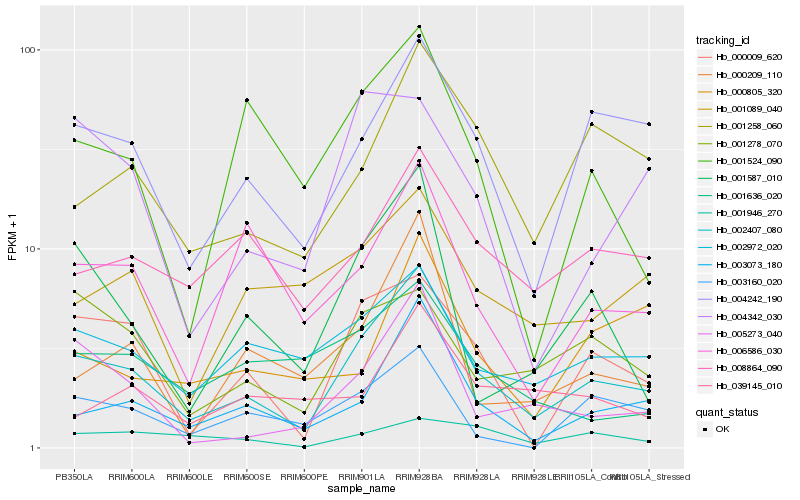
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002407_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 2 |
Hb_004242_190 |
0.1624870201 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 3 |
Hb_003073_180 |
0.167440373 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 4 |
Hb_002972_020 |
0.1835016283 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 5 |
Hb_001258_060 |
0.1863998521 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 6 |
Hb_000009_620 |
0.1886202808 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_005273_040 |
0.196630026 |
- |
- |
- |
| 8 |
Hb_008864_090 |
0.1986488448 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 9 |
Hb_001587_010 |
0.2011046451 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 10 |
Hb_006586_030 |
0.2045514519 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 11 |
Hb_000209_110 |
0.2063507563 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 12 |
Hb_003160_020 |
0.2102669014 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 13 |
Hb_001089_040 |
0.2120955581 |
transcription factor |
TF Family: bHLH |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 14 |
Hb_001524_090 |
0.2135067 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 15 |
Hb_001278_070 |
0.2140139465 |
- |
- |
hypothetical protein glysoja_020139 [Glycine soja] |
| 16 |
Hb_001636_020 |
0.2252693731 |
- |
- |
Serine/threonine-protein kinase PBS1 [Theobroma cacao] |
| 17 |
Hb_004342_030 |
0.2266009117 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 18 |
Hb_039145_010 |
0.2286697746 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 19 |
Hb_001946_270 |
0.2293801399 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 20 |
Hb_000805_320 |
0.2312557774 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |