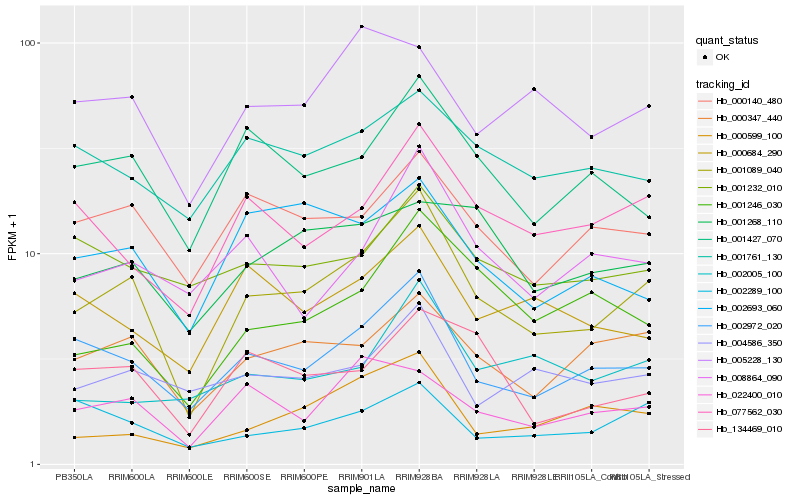
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_040 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 2 |
Hb_002972_020 |
0.119500045 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 3 |
Hb_000347_440 |
0.1373951325 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 4 |
Hb_077562_030 |
0.1534748442 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001427_070 |
0.15361667 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 6 |
Hb_000140_480 |
0.1554013899 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 7 |
Hb_001246_030 |
0.1636760823 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 8 |
Hb_022400_010 |
0.1637442838 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |
| 9 |
Hb_000599_100 |
0.165014339 |
- |
- |
hypothetical protein B456_001G254000 [Gossypium raimondii] |
| 10 |
Hb_134469_010 |
0.165599769 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008864_090 |
0.1664928795 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 12 |
Hb_004586_350 |
0.1677303389 |
- |
- |
Phosphoribosylaminoimidazole-succinocarboxamide synthase [Morus notabilis] |
| 13 |
Hb_002693_060 |
0.1692629534 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001761_130 |
0.1702235057 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002289_100 |
0.171827053 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 16 |
Hb_005228_130 |
0.172806949 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_001268_110 |
0.1734514271 |
- |
- |
PREDICTED: uncharacterized protein LOC105630183 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000684_290 |
0.1734845752 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 19 |
Hb_001232_010 |
0.1758478858 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 20 |
Hb_002005_100 |
0.1781999415 |
- |
- |
- |