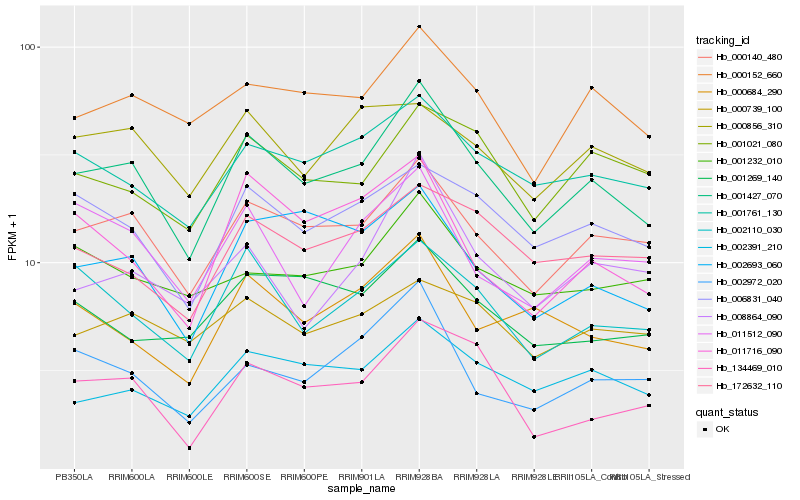
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_070 |
0.0 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 2 |
Hb_000140_480 |
0.0711553875 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 3 |
Hb_001761_130 |
0.099328178 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000152_660 |
0.0998937121 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 5 |
Hb_006831_040 |
0.1002946896 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002391_210 |
0.1017427316 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 7 |
Hb_002110_030 |
0.1077572267 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_002693_060 |
0.1110869124 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_172632_110 |
0.1120115568 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 10 |
Hb_134469_010 |
0.1124543434 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001021_080 |
0.1134072292 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 12 |
Hb_000684_290 |
0.1153530458 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 13 |
Hb_002972_020 |
0.1184480427 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 14 |
Hb_011716_090 |
0.1193845572 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 15 |
Hb_001232_010 |
0.1197936188 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 16 |
Hb_008864_090 |
0.1210462342 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 17 |
Hb_001269_140 |
0.121154315 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 18 |
Hb_000739_100 |
0.1211722489 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000856_310 |
0.121947138 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 20 |
Hb_011512_090 |
0.1220740083 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |