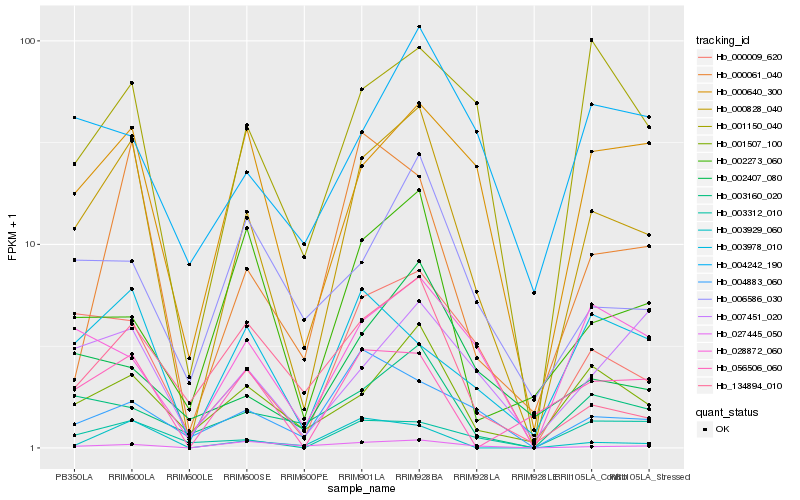
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000828_040 |
0.0 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 2 |
Hb_000009_620 |
0.1828013012 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_001507_100 |
0.200868492 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000640_300 |
0.2162054951 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 5 |
Hb_003160_020 |
0.231854327 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 6 |
Hb_028872_060 |
0.235700803 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 7 |
Hb_001150_040 |
0.2364289102 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 8 |
Hb_000061_040 |
0.2400847243 |
- |
- |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 9 |
Hb_003978_010 |
0.2430162694 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 10 |
Hb_007451_020 |
0.2432916092 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 11 |
Hb_056506_060 |
0.243940298 |
- |
- |
hypothetical protein B456_007G294000 [Gossypium raimondii] |
| 12 |
Hb_003929_060 |
0.2486076419 |
- |
- |
PREDICTED: uncharacterized protein ECU03_1610 [Jatropha curcas] |
| 13 |
Hb_134894_010 |
0.2489047463 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 14 |
Hb_003312_010 |
0.2503870138 |
- |
- |
PREDICTED: uncharacterized protein LOC104428797 [Eucalyptus grandis] |
| 15 |
Hb_027445_050 |
0.2512551629 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 16 |
Hb_002407_080 |
0.2526008851 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 17 |
Hb_006586_030 |
0.2529728864 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 18 |
Hb_004883_060 |
0.2571030981 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002273_060 |
0.2586573715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004242_190 |
0.2596033558 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |