| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
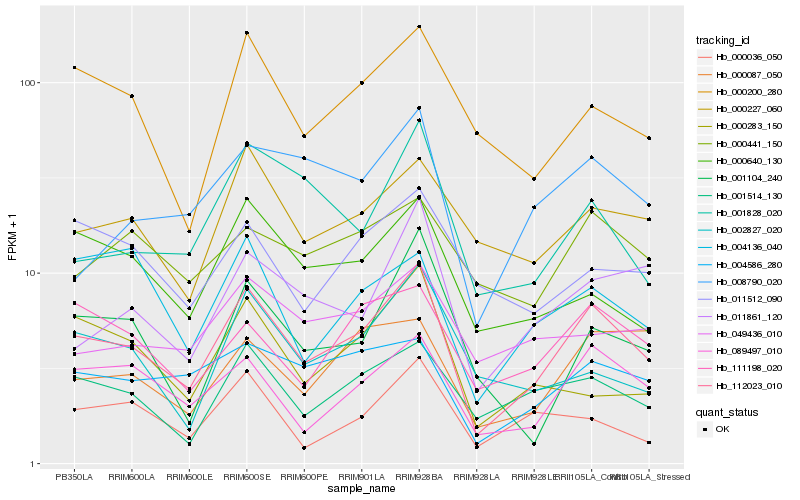
Hb_112023_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 2 |
Hb_089497_010 |
0.135975016 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_001514_130 |
0.1477908866 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 4 |
Hb_004586_280 |
0.1565204341 |
- |
- |
PREDICTED: GEM-like protein 1 [Jatropha curcas] |
| 5 |
Hb_111198_020 |
0.1615223724 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 6 |
Hb_000227_060 |
0.1618336961 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 7 |
Hb_000200_280 |
0.1622972268 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 8 |
Hb_011861_120 |
0.163193882 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 9 |
Hb_000640_130 |
0.1674975669 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000087_050 |
0.1702205715 |
- |
- |
hypothetical protein JCGZ_16227 [Jatropha curcas] |
| 11 |
Hb_001828_020 |
0.1717613206 |
- |
- |
hypothetical protein CISIN_1g010132mg [Citrus sinensis] |
| 12 |
Hb_008790_020 |
0.1735693765 |
- |
- |
PREDICTED: uncharacterized protein LOC105638122 [Jatropha curcas] |
| 13 |
Hb_000036_050 |
0.1745470114 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 14 |
Hb_049436_010 |
0.1760563165 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 15 |
Hb_002827_020 |
0.1766545627 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 16 |
Hb_001104_240 |
0.1771226171 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_004136_040 |
0.1771763739 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 18 |
Hb_000441_150 |
0.1771845173 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_011512_090 |
0.1797719392 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 20 |
Hb_000283_150 |
0.1802179546 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |