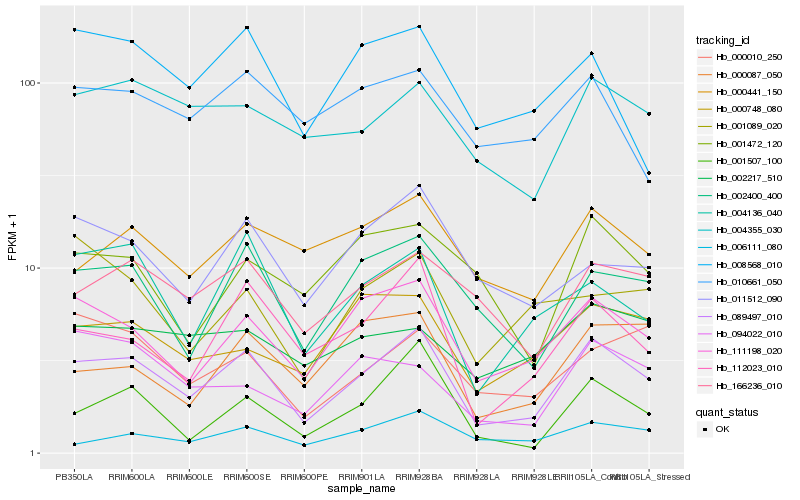
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089497_010 |
0.0 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_111198_020 |
0.1225857305 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 3 |
Hb_112023_010 |
0.135975016 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 4 |
Hb_002400_400 |
0.1380497156 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_004136_040 |
0.1485412496 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 6 |
Hb_166236_010 |
0.153054862 |
- |
- |
- |
| 7 |
Hb_004355_030 |
0.1581244989 |
- |
- |
fumarylacetoacetate hydrolase, putative [Ricinus communis] |
| 8 |
Hb_011512_090 |
0.1583678338 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 9 |
Hb_000441_150 |
0.1614752701 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_000748_080 |
0.1651645889 |
- |
- |
hypothetical protein OsI_28173 [Oryza sativa Indica Group] |
| 11 |
Hb_001507_100 |
0.1658322632 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_006111_080 |
0.1660115502 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 13 |
Hb_008568_010 |
0.1675988522 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 14 |
Hb_000087_050 |
0.1698322916 |
- |
- |
hypothetical protein JCGZ_16227 [Jatropha curcas] |
| 15 |
Hb_000010_250 |
0.1722927179 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 16 |
Hb_001472_120 |
0.1725023148 |
- |
- |
- |
| 17 |
Hb_010661_050 |
0.1764258188 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP]-like [Fragaria vesca subsp. vesca] |
| 18 |
Hb_002217_510 |
0.1778550219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_094022_010 |
0.1792215084 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 20 |
Hb_001089_020 |
0.181346026 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |