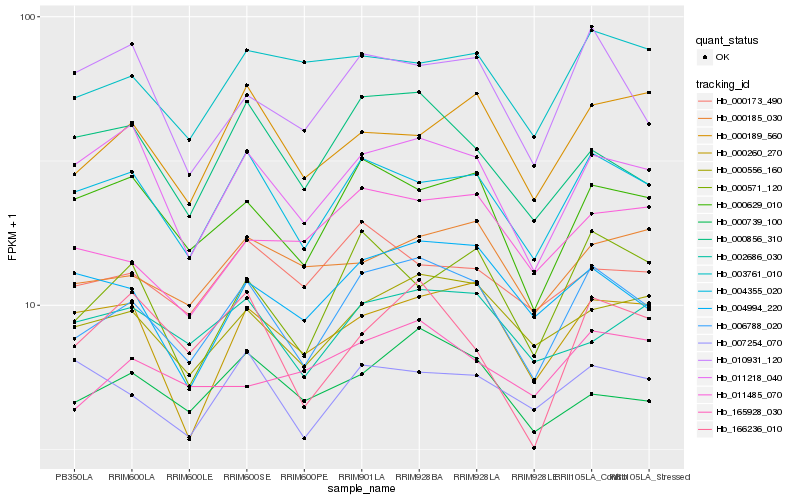
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006788_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 2 |
Hb_004355_020 |
0.0671767685 |
- |
- |
PREDICTED: lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000556_160 |
0.0772644596 |
- |
- |
PREDICTED: ankyrin repeat and zinc finger domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_000629_010 |
0.081030578 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011218_040 |
0.0833563033 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000739_100 |
0.0896659033 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 7 |
Hb_000571_120 |
0.0897897293 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 8 |
Hb_000856_310 |
0.0922239412 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 9 |
Hb_004994_220 |
0.0932055112 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_003761_010 |
0.096208211 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_010931_120 |
0.0966282987 |
- |
- |
myosin heavy chain-related family protein [Populus trichocarpa] |
| 12 |
Hb_000189_560 |
0.0968840335 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 13 |
Hb_000260_270 |
0.0976290708 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000173_490 |
0.0980868331 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000185_030 |
0.0981163807 |
- |
- |
PREDICTED: arginine-glutamic acid dipeptide repeats protein-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_002686_030 |
0.0984860149 |
- |
- |
PREDICTED: uncharacterized protein LOC105632635 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011485_070 |
0.0990115235 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 18 |
Hb_007254_070 |
0.0994681427 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_166236_010 |
0.1000274314 |
- |
- |
- |
| 20 |
Hb_165928_030 |
0.1007534751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |