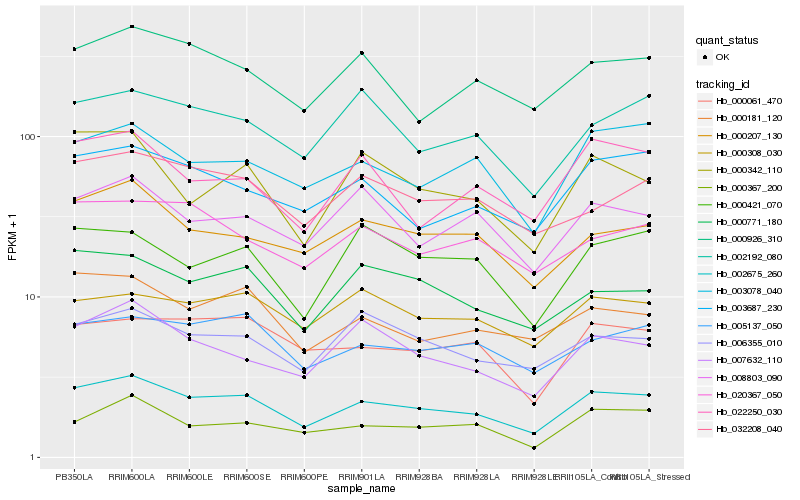
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002675_260 |
0.0 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 2 |
Hb_000207_130 |
0.0876432825 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 3 |
Hb_005137_050 |
0.0880624178 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 4 |
Hb_003687_230 |
0.0894162673 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 5 |
Hb_003078_040 |
0.0913474241 |
- |
- |
PREDICTED: hit family protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007632_110 |
0.0919921154 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 7 |
Hb_000771_180 |
0.0936339959 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 8 |
Hb_032208_040 |
0.0936809229 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 9 |
Hb_006355_010 |
0.0938639401 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 10 |
Hb_000181_120 |
0.0941469001 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 11 |
Hb_000926_310 |
0.0946221983 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 12 |
Hb_000061_470 |
0.0946613015 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 13 |
Hb_000367_200 |
0.0973417166 |
- |
- |
hypothetical protein CISIN_1g0362003mg, partial [Citrus sinensis] |
| 14 |
Hb_002192_080 |
0.097934222 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 15 |
Hb_020367_050 |
0.0983936285 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_000342_110 |
0.0991536939 |
- |
- |
PREDICTED: uncharacterized protein LOC105638174 [Jatropha curcas] |
| 17 |
Hb_000421_070 |
0.099347958 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_022250_030 |
0.0996291239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008803_090 |
0.1001753367 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |
| 20 |
Hb_000308_030 |
0.1004072753 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |