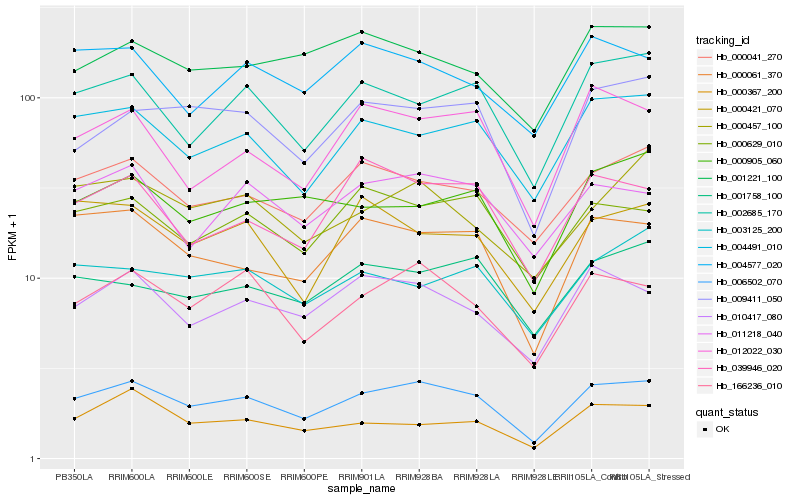
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006502_070 |
0.0 |
- |
- |
separase, putative [Ricinus communis] |
| 2 |
Hb_166236_010 |
0.0781107017 |
- |
- |
- |
| 3 |
Hb_000061_370 |
0.0856662449 |
- |
- |
PREDICTED: zinc finger RNA-binding protein [Vitis vinifera] |
| 4 |
Hb_004491_010 |
0.0883740231 |
- |
- |
60S ribosomal protein L51, putative [Ricinus communis] |
| 5 |
Hb_000041_270 |
0.0884449993 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 6 |
Hb_009411_050 |
0.0895444157 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 7 |
Hb_010417_080 |
0.0898790228 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002685_170 |
0.0935018319 |
- |
- |
hypothetical protein EUTSA_v10019281mg [Eutrema salsugineum] |
| 9 |
Hb_000629_010 |
0.0948875103 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011218_040 |
0.0969702258 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_012022_030 |
0.1021127712 |
transcription factor |
TF Family: Alfin-like |
phd/F-box containing protein, putative [Ricinus communis] |
| 12 |
Hb_039946_020 |
0.1022028471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001758_100 |
0.10395158 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000905_060 |
0.1060407232 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_000457_100 |
0.1079190539 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 16 |
Hb_004577_020 |
0.1079279382 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 17 |
Hb_001221_100 |
0.1086179469 |
- |
- |
PREDICTED: elongation factor 1-delta 1-like [Citrus sinensis] |
| 18 |
Hb_000421_070 |
0.1089288337 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_000367_200 |
0.1093915678 |
- |
- |
hypothetical protein CISIN_1g0362003mg, partial [Citrus sinensis] |
| 20 |
Hb_003125_200 |
0.1107705714 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |