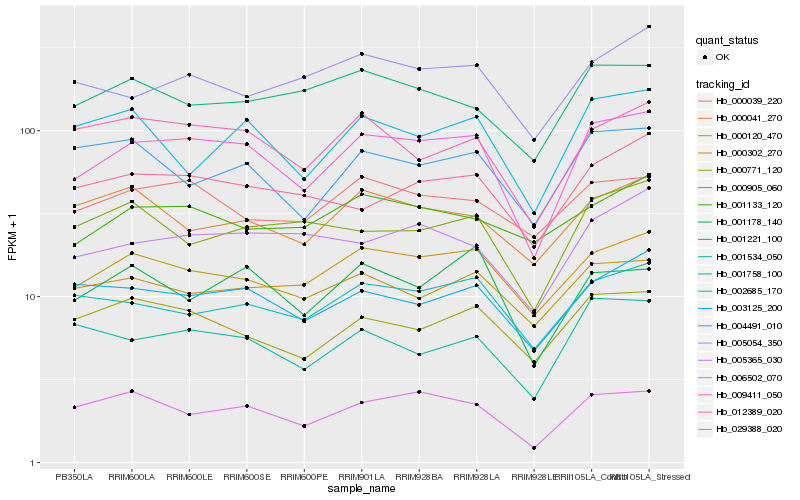
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009411_050 |
0.0 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 2 |
Hb_006502_070 |
0.0895444157 |
- |
- |
separase, putative [Ricinus communis] |
| 3 |
Hb_001758_100 |
0.0990826404 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 4 |
Hb_003125_200 |
0.0999649552 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 5 |
Hb_012389_020 |
0.1004989133 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 6 |
Hb_000771_120 |
0.1035699649 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001178_140 |
0.1057256598 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005365_030 |
0.1058204737 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 9 |
Hb_001534_050 |
0.1080424999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642299 [Jatropha curcas] |
| 10 |
Hb_000302_270 |
0.1085348639 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000039_220 |
0.1086508661 |
- |
- |
PREDICTED: vesicle-associated protein 4-2-like [Gossypium raimondii] |
| 12 |
Hb_029388_020 |
0.1089348494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000120_470 |
0.1106838218 |
- |
- |
hypothetical protein VITISV_017372 [Vitis vinifera] |
| 14 |
Hb_001221_100 |
0.1132719292 |
- |
- |
PREDICTED: elongation factor 1-delta 1-like [Citrus sinensis] |
| 15 |
Hb_002685_170 |
0.1133300276 |
- |
- |
hypothetical protein EUTSA_v10019281mg [Eutrema salsugineum] |
| 16 |
Hb_004491_010 |
0.1139143412 |
- |
- |
60S ribosomal protein L51, putative [Ricinus communis] |
| 17 |
Hb_005054_350 |
0.1143664361 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P0 [Vitis vinifera] |
| 18 |
Hb_001133_120 |
0.1153887159 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 19 |
Hb_000041_270 |
0.1157865 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 20 |
Hb_000905_060 |
0.1158992026 |
- |
- |
protein with unknown function [Ricinus communis] |