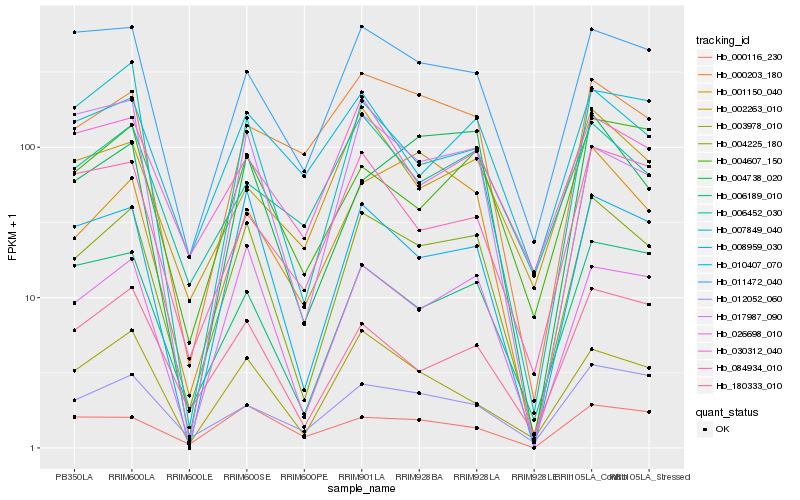
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004225_180 |
0.0 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 2 |
Hb_026698_010 |
0.107380621 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_010407_070 |
0.1150927821 |
- |
- |
hypothetical protein POPTR_0008s16750g [Populus trichocarpa] |
| 4 |
Hb_006189_010 |
0.1392070582 |
- |
- |
- |
| 5 |
Hb_011472_040 |
0.1469609505 |
- |
- |
PREDICTED: clathrin light chain 3 [Jatropha curcas] |
| 6 |
Hb_007849_040 |
0.1472837587 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 7 |
Hb_003978_010 |
0.1474712717 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 8 |
Hb_180333_010 |
0.1537110127 |
- |
- |
- |
| 9 |
Hb_004738_020 |
0.154359442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004607_150 |
0.1552084175 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 11 |
Hb_012052_060 |
0.1565396135 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |
| 12 |
Hb_001150_040 |
0.1627448194 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 13 |
Hb_000203_180 |
0.1634182195 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10 [Jatropha curcas] |
| 14 |
Hb_002263_010 |
0.1666243459 |
- |
- |
hypothetical protein CICLE_v10012259mg [Citrus clementina] |
| 15 |
Hb_006452_030 |
0.1674300026 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 16 |
Hb_084934_010 |
0.17096191 |
- |
- |
hypothetical protein JCGZ_22225 [Jatropha curcas] |
| 17 |
Hb_000116_230 |
0.171173329 |
- |
- |
- |
| 18 |
Hb_030312_040 |
0.171702107 |
- |
- |
hypothetical protein JCGZ_04876 [Jatropha curcas] |
| 19 |
Hb_017987_090 |
0.1717758011 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 2 [Jatropha curcas] |
| 20 |
Hb_008959_030 |
0.1730083741 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |