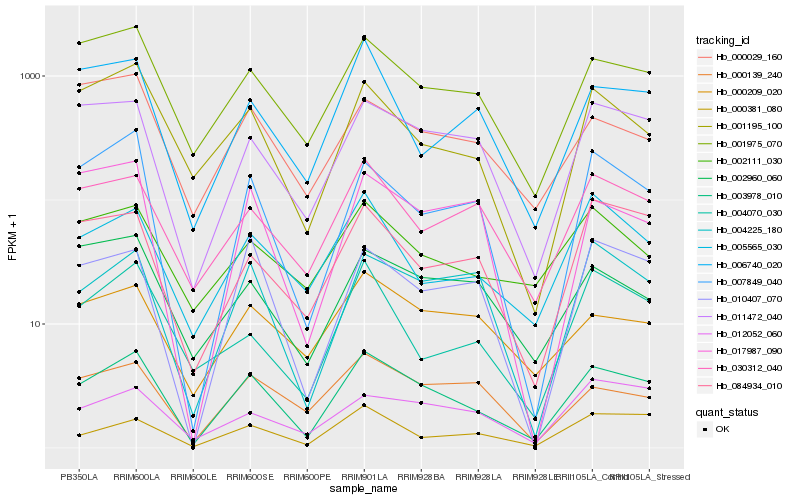
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003978_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_011472_040 |
0.134059438 |
- |
- |
PREDICTED: clathrin light chain 3 [Jatropha curcas] |
| 3 |
Hb_007849_040 |
0.1387670493 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 4 |
Hb_001975_070 |
0.1393340169 |
- |
- |
eukaryotic translation elongation factor 1B alpha-subunit [Hevea brasiliensis] |
| 5 |
Hb_004225_180 |
0.1474712717 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 6 |
Hb_001195_100 |
0.1497363075 |
- |
- |
PREDICTED: uncharacterized protein LOC105633812 [Jatropha curcas] |
| 7 |
Hb_010407_070 |
0.1560937957 |
- |
- |
hypothetical protein POPTR_0008s16750g [Populus trichocarpa] |
| 8 |
Hb_084934_010 |
0.1561799172 |
- |
- |
hypothetical protein JCGZ_22225 [Jatropha curcas] |
| 9 |
Hb_005565_030 |
0.1592504825 |
- |
- |
PREDICTED: uncharacterized protein LOC105633107 isoform X1 [Jatropha curcas] |
| 10 |
Hb_030312_040 |
0.1595493912 |
- |
- |
hypothetical protein JCGZ_04876 [Jatropha curcas] |
| 11 |
Hb_000381_080 |
0.1618380807 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair protein RAD16 [Jatropha curcas] |
| 12 |
Hb_000139_240 |
0.1653512738 |
- |
- |
PREDICTED: putative F-box protein At3g16210 [Jatropha curcas] |
| 13 |
Hb_000209_020 |
0.1681331579 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 14 |
Hb_017987_090 |
0.1683539718 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 2 [Jatropha curcas] |
| 15 |
Hb_006740_020 |
0.1702339714 |
- |
- |
PREDICTED: bis(5'-adenosyl)-triphosphatase [Jatropha curcas] |
| 16 |
Hb_002960_060 |
0.1711498894 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 28 [Jatropha curcas] |
| 17 |
Hb_012052_060 |
0.1714000359 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |
| 18 |
Hb_002111_030 |
0.1747974887 |
- |
- |
PREDICTED: NEDD8 ultimate buster 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000029_160 |
0.1758952315 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 20 |
Hb_004070_030 |
0.1763372973 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase, mitochondrial isoform X2 [Jatropha curcas] |