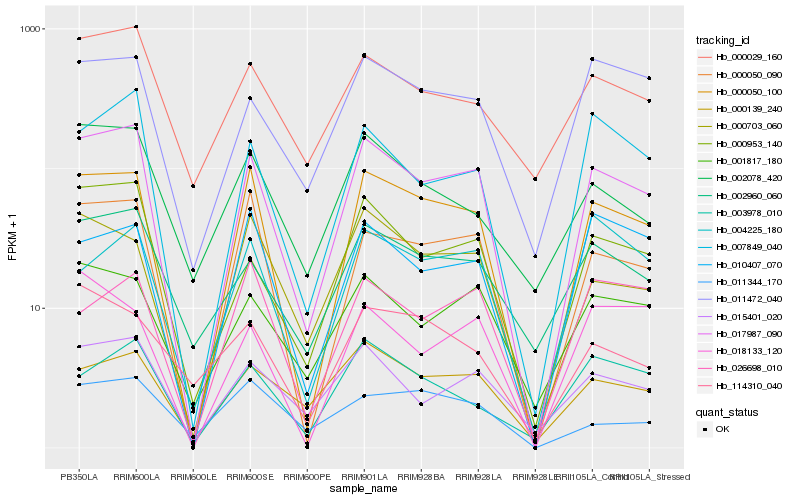
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000050_100 |
0.0 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 2 [Jatropha curcas] |
| 2 |
Hb_017987_090 |
0.0926332729 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 2 [Jatropha curcas] |
| 3 |
Hb_000050_090 |
0.0972628318 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 4 |
Hb_000703_060 |
0.1347476711 |
- |
- |
phosphoenolpyruvate carboxylase [Manihot esculenta] |
| 5 |
Hb_010407_070 |
0.144914747 |
- |
- |
hypothetical protein POPTR_0008s16750g [Populus trichocarpa] |
| 6 |
Hb_026698_010 |
0.1623050796 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_000139_240 |
0.1744023473 |
- |
- |
PREDICTED: putative F-box protein At3g16210 [Jatropha curcas] |
| 8 |
Hb_004225_180 |
0.1765904445 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 9 |
Hb_003978_010 |
0.1776014581 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 10 |
Hb_007849_040 |
0.178403279 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 11 |
Hb_114310_040 |
0.182830125 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 12 |
Hb_001817_180 |
0.1836549892 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 13 |
Hb_011472_040 |
0.1845394077 |
- |
- |
PREDICTED: clathrin light chain 3 [Jatropha curcas] |
| 14 |
Hb_002078_420 |
0.1860775127 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 15 |
Hb_015401_020 |
0.1888577839 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 16 |
Hb_000953_140 |
0.1890923851 |
transcription factor |
TF Family: Trihelix |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 17 |
Hb_011344_170 |
0.1891597271 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 4-like, partial [Camelina sativa] |
| 18 |
Hb_002960_060 |
0.1917327654 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 28 [Jatropha curcas] |
| 19 |
Hb_000029_160 |
0.1931671429 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 20 |
Hb_018133_120 |
0.1937462874 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |