| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
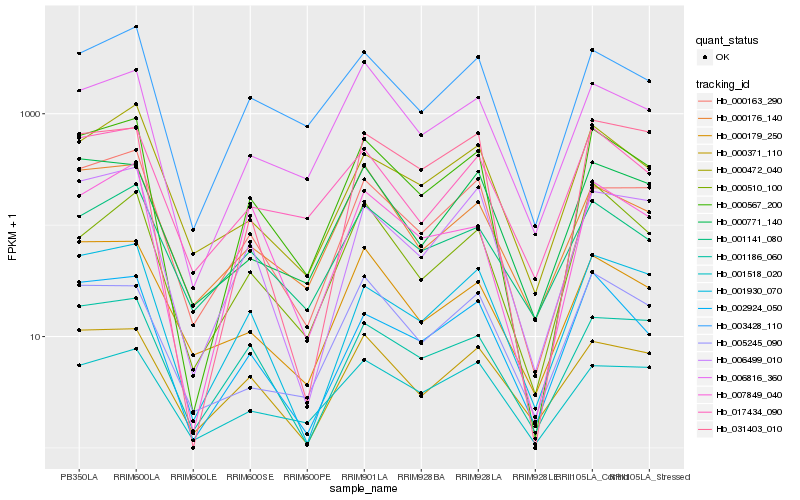
Hb_000567_200 |
0.0 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10 [Jatropha curcas] |
| 2 |
Hb_003428_110 |
0.1023449854 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 3 |
Hb_000163_290 |
0.110259969 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |
| 4 |
Hb_002924_050 |
0.1114731202 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 5 |
Hb_006499_010 |
0.1167532639 |
- |
- |
- |
| 6 |
Hb_001930_070 |
0.1175056227 |
- |
- |
- |
| 7 |
Hb_001518_020 |
0.1209339709 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 13-like isoform X2 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_006816_360 |
0.1257971525 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 3 [Jatropha curcas] |
| 9 |
Hb_017434_090 |
0.1262592025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000472_040 |
0.1276904446 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 11 |
Hb_000371_110 |
0.128230117 |
- |
- |
- |
| 12 |
Hb_007849_040 |
0.1311546803 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 13 |
Hb_001186_060 |
0.1314528559 |
- |
- |
- |
| 14 |
Hb_000176_140 |
0.131689067 |
- |
- |
PREDICTED: bystin [Jatropha curcas] |
| 15 |
Hb_000771_140 |
0.1317043767 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 16 |
Hb_000179_250 |
0.1331220575 |
- |
- |
PREDICTED: WD repeat-containing protein 61-like [Citrus sinensis] |
| 17 |
Hb_000510_100 |
0.1349851078 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_031403_010 |
0.1359438873 |
- |
- |
hypothetical protein POPTR_0001s40140g [Populus trichocarpa] |
| 19 |
Hb_005245_090 |
0.1369559631 |
- |
- |
PREDICTED: peroxisomal membrane protein 13 [Jatropha curcas] |
| 20 |
Hb_001141_080 |
0.1370676639 |
- |
- |
PREDICTED: protein RDM1 [Jatropha curcas] |