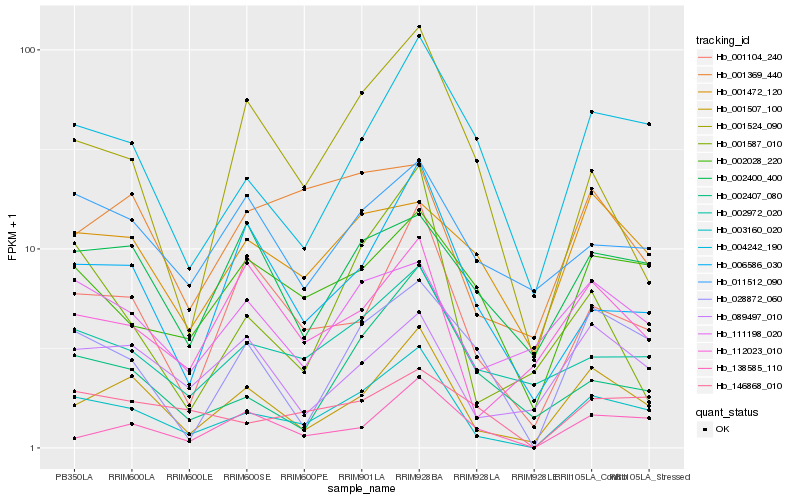
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003160_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 2 |
Hb_001507_100 |
0.153759729 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_004242_190 |
0.176805666 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_001104_240 |
0.1773701898 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_002972_020 |
0.1858754342 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 6 |
Hb_002028_220 |
0.1948448033 |
- |
- |
aminoadipic semialdehyde synthase, putative [Ricinus communis] |
| 7 |
Hb_028872_060 |
0.1960177291 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 8 |
Hb_006586_030 |
0.2005983182 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 9 |
Hb_146868_010 |
0.205352423 |
- |
- |
- |
| 10 |
Hb_138585_110 |
0.2091149799 |
- |
- |
- |
| 11 |
Hb_002407_080 |
0.2102669014 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 12 |
Hb_111198_020 |
0.2130287338 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 13 |
Hb_089497_010 |
0.2149742703 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_112023_010 |
0.2154839271 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 15 |
Hb_001472_120 |
0.2221334596 |
- |
- |
- |
| 16 |
Hb_011512_090 |
0.2235472883 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 17 |
Hb_002400_400 |
0.2237241384 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_001524_090 |
0.2241415306 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 19 |
Hb_001369_440 |
0.2243124987 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 20 |
Hb_001587_010 |
0.2250127778 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |