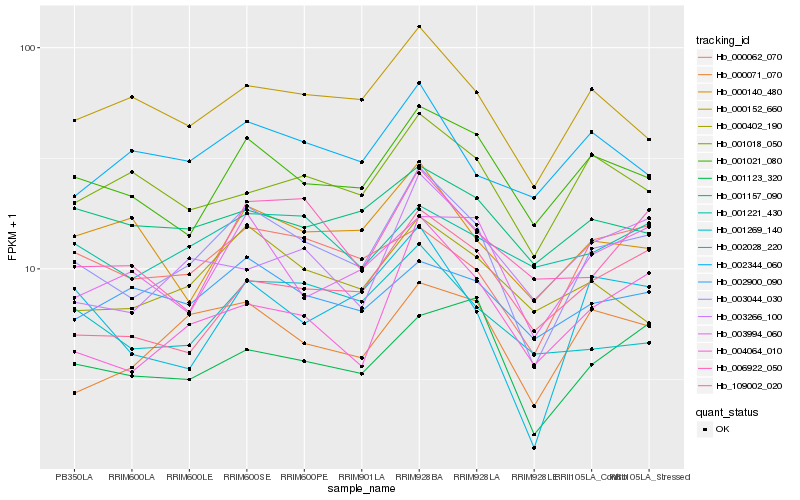
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003044_030 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Cicer arietinum] |
| 2 |
Hb_001021_080 |
0.1092245342 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 3 |
Hb_000071_070 |
0.1164370379 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 4 |
Hb_000152_660 |
0.1194905228 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 5 |
Hb_004064_010 |
0.1204156465 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 6 |
Hb_002028_220 |
0.1264479588 |
- |
- |
aminoadipic semialdehyde synthase, putative [Ricinus communis] |
| 7 |
Hb_000402_190 |
0.127186267 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 8 |
Hb_000062_070 |
0.1278159769 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 9 |
Hb_001123_320 |
0.1281776737 |
- |
- |
PREDICTED: uncharacterized protein LOC105647314 isoform X3 [Jatropha curcas] |
| 10 |
Hb_109002_020 |
0.1285954531 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 11 |
Hb_003994_060 |
0.1313740328 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 12 |
Hb_001269_140 |
0.13169436 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 13 |
Hb_003266_100 |
0.1332748984 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 14 |
Hb_001018_050 |
0.1345634734 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 15 |
Hb_006922_050 |
0.1350824938 |
- |
- |
PREDICTED: dipeptidyl peptidase 8 [Jatropha curcas] |
| 16 |
Hb_002900_090 |
0.1352408505 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 17 |
Hb_001157_090 |
0.1365167244 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 18 |
Hb_000140_480 |
0.1370864934 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 19 |
Hb_001221_430 |
0.1385695393 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At5g63520 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002344_060 |
0.1405982363 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |