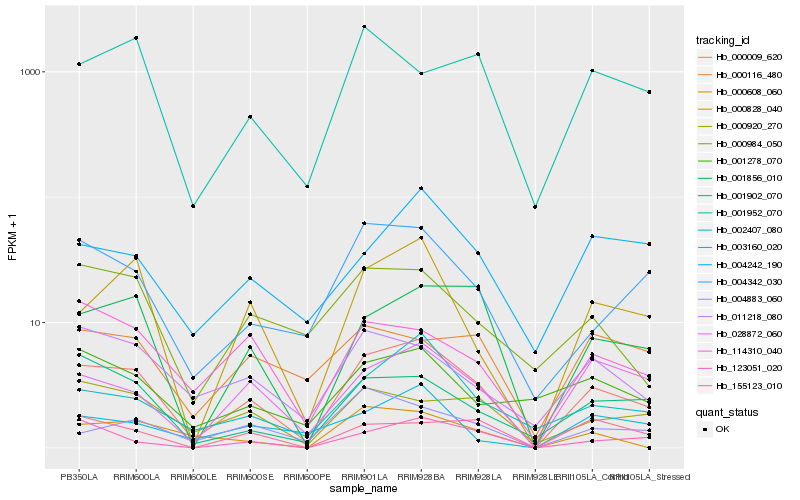
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_620 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_001856_010 |
0.1823764157 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_028872_060 |
0.1826624693 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 4 |
Hb_000828_040 |
0.1828013012 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 5 |
Hb_002407_080 |
0.1886202808 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 6 |
Hb_004342_030 |
0.1889101239 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 7 |
Hb_114310_040 |
0.2024563435 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 8 |
Hb_000920_270 |
0.2045481588 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 9 |
Hb_011218_080 |
0.205358156 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 10 |
Hb_001902_070 |
0.2053842715 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_004242_190 |
0.2079721045 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 12 |
Hb_000984_050 |
0.2104553355 |
- |
- |
sucrose transporter 5 [Hevea brasiliensis] |
| 13 |
Hb_155123_010 |
0.2165582445 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 14 |
Hb_001952_070 |
0.2168218966 |
- |
- |
40S ribosomal protein S6B [Hevea brasiliensis] |
| 15 |
Hb_123051_020 |
0.2170580239 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 16 |
Hb_001278_070 |
0.2184156771 |
- |
- |
hypothetical protein glysoja_020139 [Glycine soja] |
| 17 |
Hb_004883_060 |
0.2196792154 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003160_020 |
0.2250524931 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 19 |
Hb_000116_480 |
0.2269455111 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 20 |
Hb_000608_060 |
0.2292240697 |
- |
- |
- |