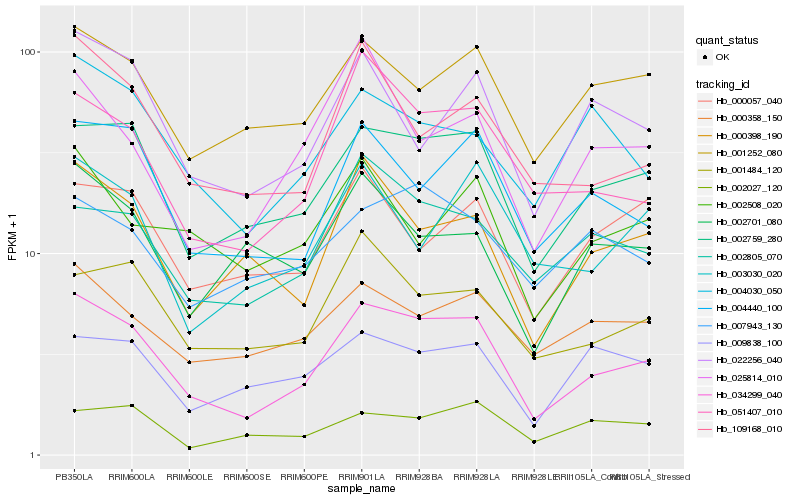
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034299_040 |
0.0 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 2 |
Hb_002759_280 |
0.0981710667 |
- |
- |
PREDICTED: uncharacterized protein LOC105649824 [Jatropha curcas] |
| 3 |
Hb_004440_100 |
0.1255017125 |
- |
- |
PREDICTED: transmembrane protein 53 [Jatropha curcas] |
| 4 |
Hb_051407_010 |
0.1279133813 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 5 |
Hb_000398_190 |
0.1357187561 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001252_080 |
0.1365970264 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit G-like [Jatropha curcas] |
| 7 |
Hb_109168_010 |
0.1380250477 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 8 |
Hb_001484_120 |
0.1386927962 |
- |
- |
hypothetical protein POPTR_2287s00200g, partial [Populus trichocarpa] |
| 9 |
Hb_002805_070 |
0.1427298426 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X2 [Jatropha curcas] |
| 10 |
Hb_022256_040 |
0.143514739 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase rbrA isoform X2 [Jatropha curcas] |
| 11 |
Hb_000358_150 |
0.1443581706 |
- |
- |
hypothetical protein VITISV_012309 [Vitis vinifera] |
| 12 |
Hb_004030_050 |
0.1507999021 |
- |
- |
PREDICTED: uncharacterized protein LOC105646691 [Jatropha curcas] |
| 13 |
Hb_000057_040 |
0.1539617575 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_003030_020 |
0.154055971 |
- |
- |
PREDICTED: uncharacterized protein LOC105632544 [Jatropha curcas] |
| 15 |
Hb_009838_100 |
0.1542556672 |
- |
- |
hypothetical protein RCOM_1217340 [Ricinus communis] |
| 16 |
Hb_025814_010 |
0.1558491847 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 17 |
Hb_002508_020 |
0.1561990759 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 18 |
Hb_002027_120 |
0.1565508322 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 19 |
Hb_002701_080 |
0.1566588523 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 20 |
Hb_007943_130 |
0.1570543366 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |