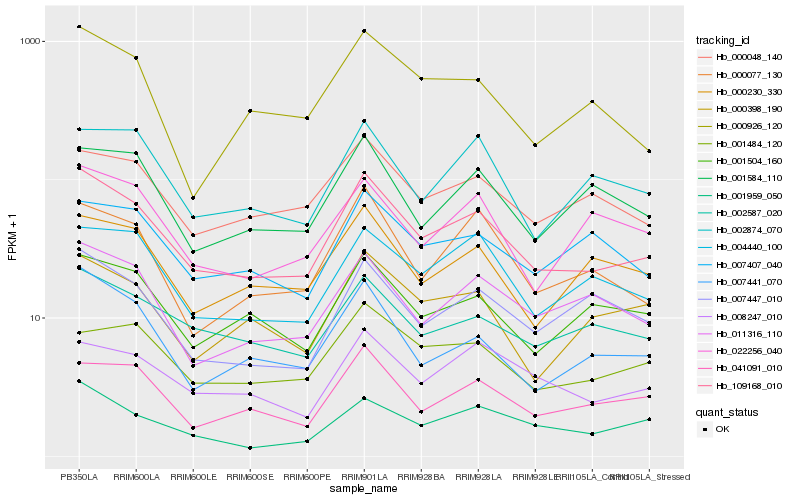
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_109168_010 |
0.0 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 2 |
Hb_007441_070 |
0.1063273138 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 3 |
Hb_002587_020 |
0.1071535277 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_011316_110 |
0.1121774073 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 5 |
Hb_001504_160 |
0.1125724306 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 6 |
Hb_041091_010 |
0.1137397813 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000048_140 |
0.1146841967 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 8 |
Hb_004440_100 |
0.1152450361 |
- |
- |
PREDICTED: transmembrane protein 53 [Jatropha curcas] |
| 9 |
Hb_001959_050 |
0.1174726964 |
- |
- |
PREDICTED: uncharacterized protein LOC103323811 isoform X1 [Prunus mume] |
| 10 |
Hb_000398_190 |
0.1186634685 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000230_330 |
0.1189197703 |
- |
- |
PREDICTED: calcium-binding protein 39-like [Populus euphratica] |
| 12 |
Hb_008247_010 |
0.1194787598 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 13 |
Hb_000077_130 |
0.1197327039 |
- |
- |
PREDICTED: callose synthase 12 [Jatropha curcas] |
| 14 |
Hb_007447_010 |
0.1200811279 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000926_120 |
0.1214184954 |
- |
- |
PREDICTED: aspartic proteinase A1-like [Jatropha curcas] |
| 16 |
Hb_002874_070 |
0.124090884 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 17 |
Hb_001584_110 |
0.124324956 |
- |
- |
PREDICTED: importin subunit alpha-4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007407_040 |
0.124895799 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_022256_040 |
0.1250681287 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase rbrA isoform X2 [Jatropha curcas] |
| 20 |
Hb_001484_120 |
0.1255034313 |
- |
- |
hypothetical protein POPTR_2287s00200g, partial [Populus trichocarpa] |