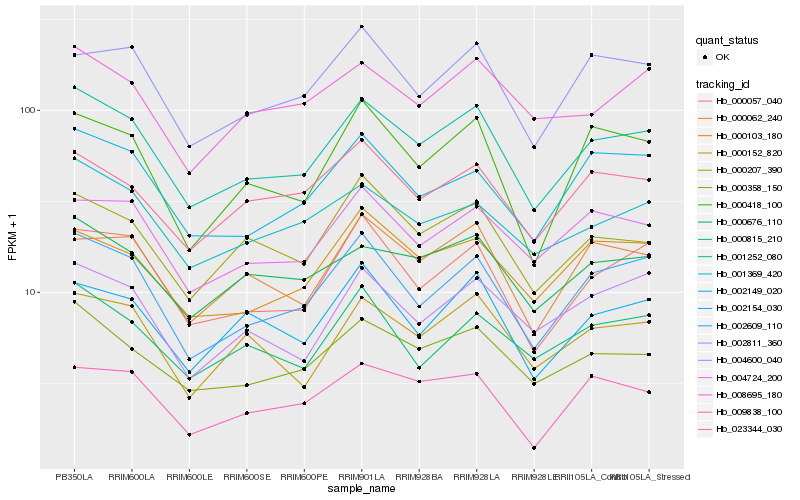
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001252_080 |
0.0 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit G-like [Jatropha curcas] |
| 2 |
Hb_000358_150 |
0.0575412466 |
- |
- |
hypothetical protein VITISV_012309 [Vitis vinifera] |
| 3 |
Hb_002609_110 |
0.0605246603 |
- |
- |
hypothetical protein JCGZ_08497 [Jatropha curcas] |
| 4 |
Hb_000676_110 |
0.0716478258 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 5 |
Hb_000057_040 |
0.0730360924 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_001369_420 |
0.0753583459 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 7 |
Hb_000207_390 |
0.0765837198 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 8 |
Hb_004724_200 |
0.078943157 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 9 |
Hb_000152_820 |
0.079388857 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g64320, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002149_020 |
0.0802963729 |
- |
- |
PREDICTED: uncharacterized protein LOC105647982 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000418_100 |
0.0818447884 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 12 |
Hb_000062_240 |
0.0829122549 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 13 |
Hb_000103_180 |
0.0829275768 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 14 |
Hb_002811_360 |
0.0833622897 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 15 |
Hb_023344_030 |
0.0846363176 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 16 |
Hb_009838_100 |
0.0855606674 |
- |
- |
hypothetical protein RCOM_1217340 [Ricinus communis] |
| 17 |
Hb_002154_030 |
0.0857037535 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_004600_040 |
0.0864649818 |
- |
- |
- |
| 19 |
Hb_008695_180 |
0.0870335365 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 20 |
Hb_000815_210 |
0.0882765207 |
- |
- |
uncoupling protein 3, partial [Manihot esculenta] |