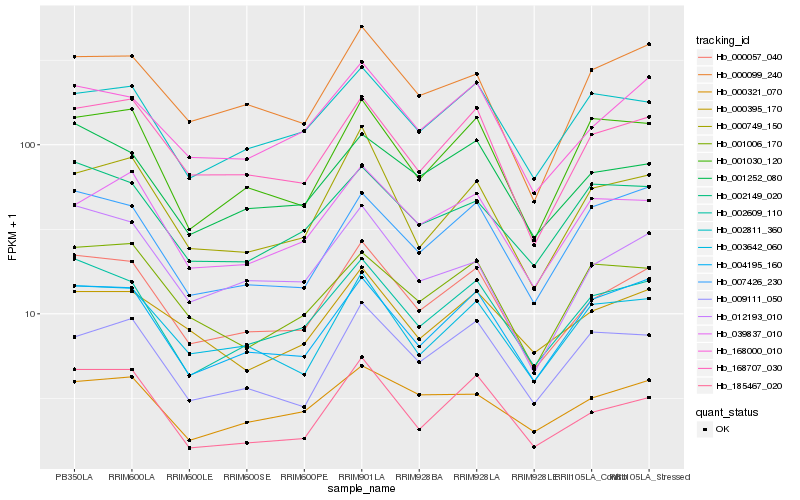
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000057_040 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_002609_110 |
0.0571085624 |
- |
- |
hypothetical protein JCGZ_08497 [Jatropha curcas] |
| 3 |
Hb_168707_030 |
0.0579902289 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 4 |
Hb_168000_010 |
0.0620575859 |
- |
- |
Calcium-dependent lipid-binding family protein isoform 2 [Theobroma cacao] |
| 5 |
Hb_003642_060 |
0.0687280464 |
- |
- |
hypothetical protein RCOM_1189860 [Ricinus communis] |
| 6 |
Hb_004195_160 |
0.0692886208 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 7 |
Hb_001252_080 |
0.0730360924 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit G-like [Jatropha curcas] |
| 8 |
Hb_000099_240 |
0.0781513097 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 9 |
Hb_012193_010 |
0.0811908486 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_000749_150 |
0.0820637816 |
- |
- |
- |
| 11 |
Hb_001030_120 |
0.083354352 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 12 |
Hb_185467_020 |
0.0849023446 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 13 |
Hb_002811_360 |
0.0852263455 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 14 |
Hb_007426_230 |
0.0856843934 |
- |
- |
hypothetical protein CICLE_v10017932mg [Citrus clementina] |
| 15 |
Hb_000321_070 |
0.0860711374 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_039837_010 |
0.0867132489 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 17 |
Hb_000395_170 |
0.0871081431 |
- |
- |
PREDICTED: probable transcriptional regulatory protein At2g25830 [Jatropha curcas] |
| 18 |
Hb_001006_170 |
0.0871806835 |
- |
- |
Endonuclease V, putative [Ricinus communis] |
| 19 |
Hb_009111_050 |
0.0877620568 |
transcription factor |
TF Family: Jumonji |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540 [Populus euphratica] |
| 20 |
Hb_002149_020 |
0.0878288108 |
- |
- |
PREDICTED: uncharacterized protein LOC105647982 isoform X2 [Jatropha curcas] |