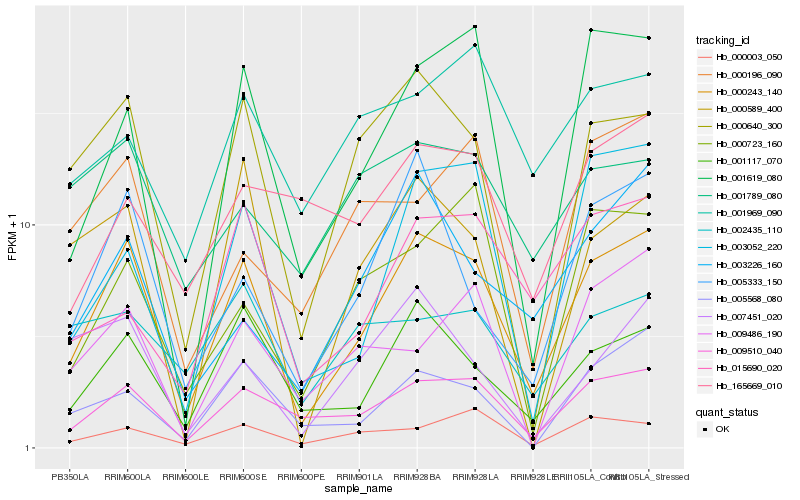
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_140 |
0.0 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 2 |
Hb_009510_040 |
0.138310012 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001117_070 |
0.1451739561 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 4 |
Hb_000640_300 |
0.1587725814 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 5 |
Hb_001619_080 |
0.1762302104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005333_150 |
0.1820050999 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 7 |
Hb_003052_220 |
0.1875849687 |
- |
- |
- |
| 8 |
Hb_005568_080 |
0.1891667578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000003_050 |
0.1894624189 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 10 |
Hb_003226_160 |
0.1983253142 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 11 |
Hb_000589_400 |
0.198646402 |
- |
- |
ESC, putative [Ricinus communis] |
| 12 |
Hb_015690_020 |
0.1998775389 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 13 |
Hb_007451_020 |
0.2049384883 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 14 |
Hb_000723_160 |
0.2101127051 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP11 [Jatropha curcas] |
| 15 |
Hb_009486_190 |
0.2113451622 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000196_090 |
0.2117339039 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 17 |
Hb_001789_080 |
0.2120998838 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 18 |
Hb_001969_090 |
0.2122757918 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 19 |
Hb_002435_110 |
0.2141693856 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 20 |
Hb_165669_010 |
0.214669196 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |