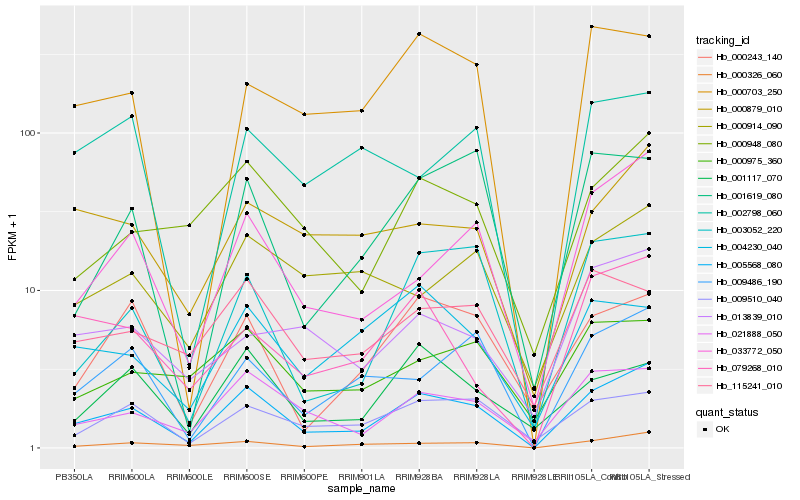
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005568_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_021888_050 |
0.1493081007 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 3 |
Hb_000326_060 |
0.1524919285 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 4 |
Hb_033772_050 |
0.1557029892 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 5 |
Hb_009486_190 |
0.1730121581 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_009510_040 |
0.1747852745 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_003052_220 |
0.17516085 |
- |
- |
- |
| 8 |
Hb_000948_080 |
0.1771532631 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 9 |
Hb_000703_250 |
0.1817276576 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10-like [Jatropha curcas] |
| 10 |
Hb_000879_010 |
0.1846794122 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 11 |
Hb_000243_140 |
0.1891667578 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 12 |
Hb_079268_010 |
0.1940101008 |
- |
- |
- |
| 13 |
Hb_001619_080 |
0.1962451821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000914_090 |
0.1983644798 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 15 |
Hb_001117_070 |
0.198607575 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 16 |
Hb_115241_010 |
0.2004101037 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 17 |
Hb_000975_360 |
0.2004483239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_013839_010 |
0.202025832 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 19 |
Hb_004230_040 |
0.203076734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002798_060 |
0.2042928138 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |