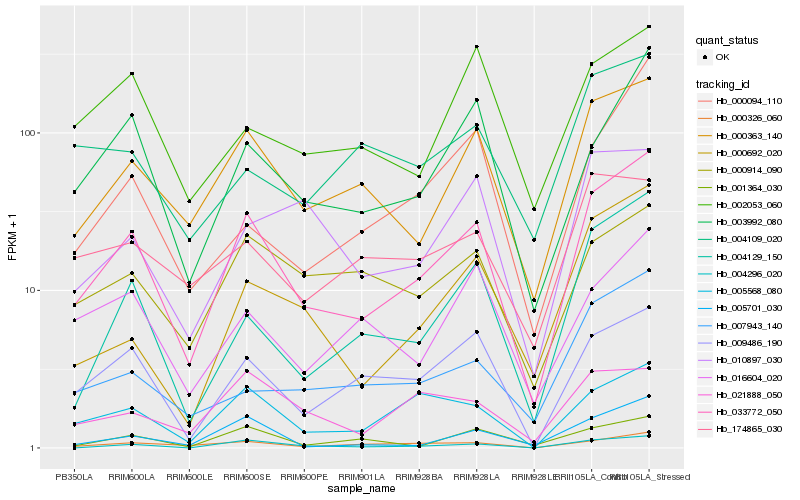
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033772_050 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 2 |
Hb_000363_140 |
0.1299816602 |
- |
- |
hypothetical protein CICLE_v10009052mg [Citrus clementina] |
| 3 |
Hb_009486_190 |
0.1513314162 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_003992_080 |
0.1535315176 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 5 |
Hb_005568_080 |
0.1557029892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000326_060 |
0.1567051788 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 7 |
Hb_004129_150 |
0.1588713023 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 8 |
Hb_005701_030 |
0.1685342666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000692_020 |
0.1726358989 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |
| 10 |
Hb_004296_020 |
0.1738282648 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_001364_030 |
0.174318785 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 12 |
Hb_016604_020 |
0.1793108108 |
- |
- |
- |
| 13 |
Hb_007943_140 |
0.183800138 |
- |
- |
PREDICTED: uncharacterized protein LOC103340977 [Prunus mume] |
| 14 |
Hb_000914_090 |
0.1857892209 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 15 |
Hb_002053_060 |
0.1884421117 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 16 |
Hb_000094_110 |
0.1907666974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010897_030 |
0.1941108805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_021888_050 |
0.1944620419 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 19 |
Hb_004109_020 |
0.1974388771 |
- |
- |
60S ribosomal protein L18aA [Hevea brasiliensis] |
| 20 |
Hb_174865_030 |
0.1978804625 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |