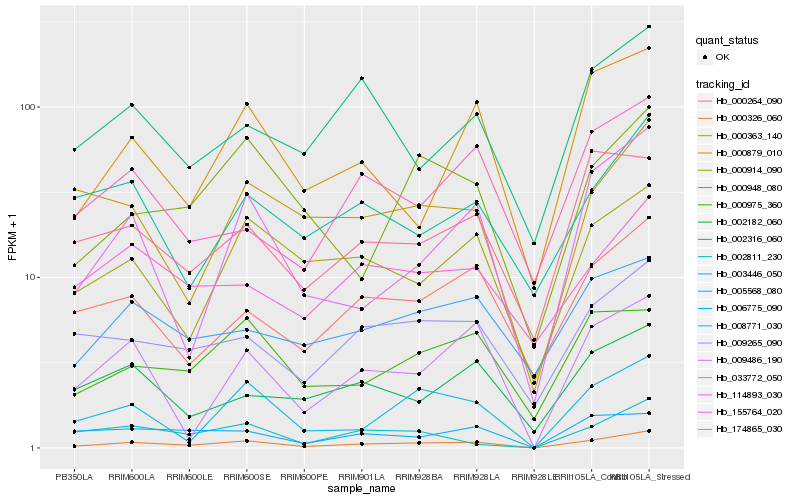
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000326_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 2 |
Hb_005568_080 |
0.1524919285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_033772_050 |
0.1567051788 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 4 |
Hb_009265_090 |
0.1652141073 |
- |
- |
PREDICTED: uncharacterized protein LOC105649618 [Jatropha curcas] |
| 5 |
Hb_009486_190 |
0.1660230358 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000264_090 |
0.1670873695 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 7 |
Hb_000363_140 |
0.168480634 |
- |
- |
hypothetical protein CICLE_v10009052mg [Citrus clementina] |
| 8 |
Hb_000914_090 |
0.1723718664 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 9 |
Hb_000948_080 |
0.1784825341 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 10 |
Hb_000975_360 |
0.1800480463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006775_090 |
0.1828443834 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 12 |
Hb_002316_060 |
0.1837609444 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 13 |
Hb_008771_030 |
0.1838237888 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 14 |
Hb_174865_030 |
0.1839883443 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |
| 15 |
Hb_000879_010 |
0.1856740151 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 16 |
Hb_155764_020 |
0.1879507499 |
- |
- |
hypothetical protein B456_009G265500 [Gossypium raimondii] |
| 17 |
Hb_003446_050 |
0.1885146009 |
- |
- |
hypothetical protein B456_002G034000, partial [Gossypium raimondii] |
| 18 |
Hb_114893_030 |
0.1897102716 |
- |
- |
PREDICTED: ribosome-recycling factor, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002182_060 |
0.1902658882 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATXR6 [Jatropha curcas] |
| 20 |
Hb_002811_230 |
0.1904414811 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |