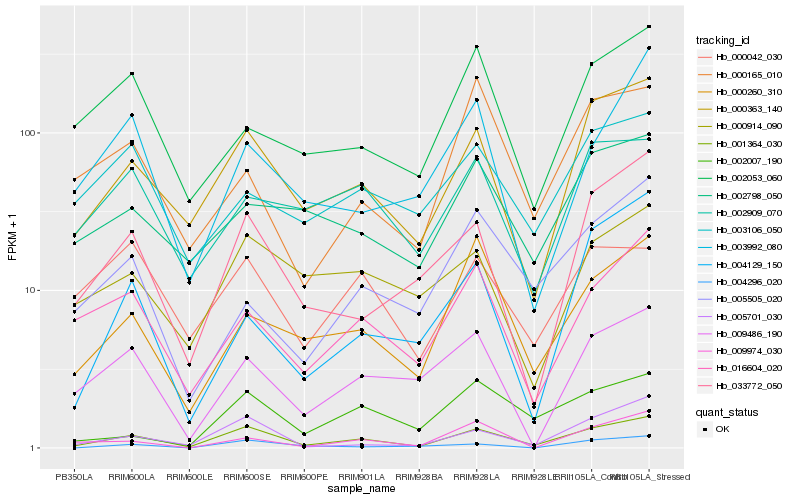
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001364_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 2 |
Hb_000363_140 |
0.1326975758 |
- |
- |
hypothetical protein CICLE_v10009052mg [Citrus clementina] |
| 3 |
Hb_005701_030 |
0.1697444038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_033772_050 |
0.174318785 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 5 |
Hb_000260_310 |
0.1878596906 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 6 |
Hb_016604_020 |
0.1892318996 |
- |
- |
- |
| 7 |
Hb_009486_190 |
0.1997688722 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_002007_190 |
0.2005359755 |
- |
- |
hypothetical protein RCOM_1432220 [Ricinus communis] |
| 9 |
Hb_000914_090 |
0.2084957768 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 10 |
Hb_004129_150 |
0.2143201798 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 11 |
Hb_002053_060 |
0.2149665175 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 12 |
Hb_002909_070 |
0.2152841153 |
- |
- |
hypothetical protein POPTR_0006s03000g [Populus trichocarpa] |
| 13 |
Hb_004296_020 |
0.2153058876 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_002798_050 |
0.2160710944 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 15 |
Hb_003992_080 |
0.2161937338 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 16 |
Hb_000165_010 |
0.2193652277 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 17 |
Hb_009974_030 |
0.219599268 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_005505_020 |
0.2212347819 |
- |
- |
Potassium channel AKT1, putative [Ricinus communis] |
| 19 |
Hb_000042_030 |
0.223270896 |
transcription factor |
TF Family: NAC |
NAC domain protein, IPR003441 [Theobroma cacao] |
| 20 |
Hb_003106_050 |
0.2250291729 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0015s08130g [Populus trichocarpa] |