| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
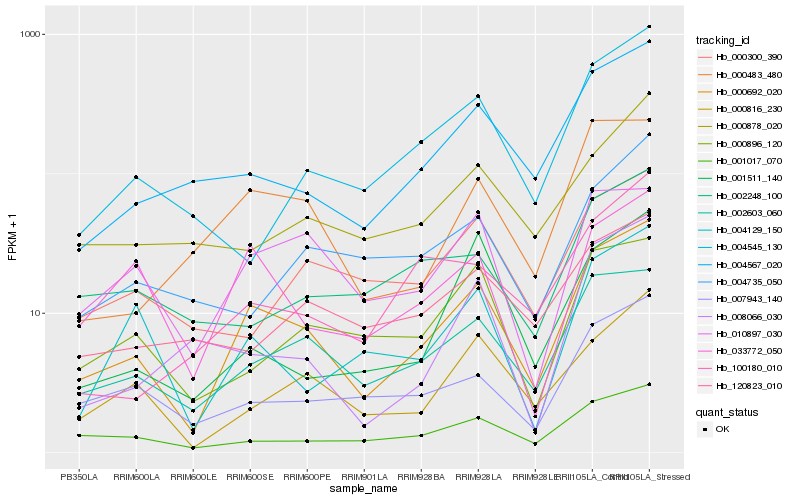
Hb_000692_020 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |
| 2 |
Hb_002603_060 |
0.1389784398 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 3 |
Hb_004567_020 |
0.160737283 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 4 |
Hb_001017_070 |
0.1642161261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000483_480 |
0.1646622486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000816_230 |
0.1663484136 |
- |
- |
PREDICTED: adenine/guanine permease AZG1 [Jatropha curcas] |
| 7 |
Hb_007943_140 |
0.1715632675 |
- |
- |
PREDICTED: uncharacterized protein LOC103340977 [Prunus mume] |
| 8 |
Hb_033772_050 |
0.1726358989 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 9 |
Hb_010897_030 |
0.1798338088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004129_150 |
0.1839718875 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 11 |
Hb_000896_120 |
0.1901053937 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 12 |
Hb_000300_390 |
0.1937569484 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 13 |
Hb_000878_020 |
0.1942646972 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 14 |
Hb_004545_130 |
0.1958152047 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 15 |
Hb_100180_010 |
0.1977100029 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 16 |
Hb_001511_140 |
0.1978746961 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 17 |
Hb_008066_030 |
0.19817158 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 18 |
Hb_120823_010 |
0.1987130281 |
- |
- |
- |
| 19 |
Hb_004735_050 |
0.2002347582 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002248_100 |
0.2009442865 |
- |
- |
hypothetical protein VITISV_027197 [Vitis vinifera] |